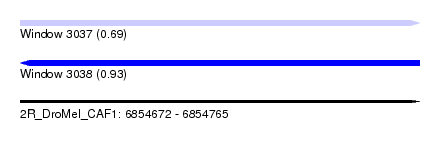
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,854,672 – 6,854,765 |
| Length | 93 |
| Max. P | 0.930157 |

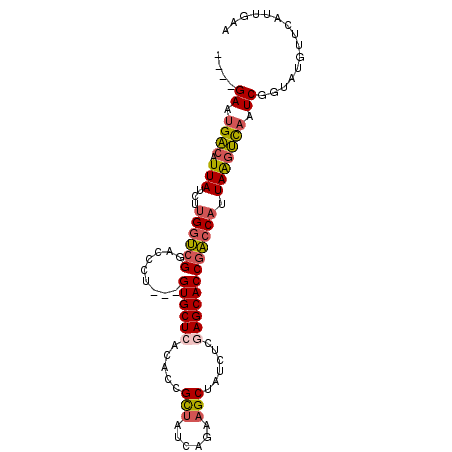
| Location | 6,854,672 – 6,854,765 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -18.40 |
| Energy contribution | -18.73 |
| Covariance contribution | 0.34 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.693761 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6854672 93 + 20766785 UUCAAUGAACAUGCCGAUUGAGUUAAUGGUCGGUGCUUAAGAUAGCUUCUGAUAGCGGUGUGAGCAC---AGGGUCCGACCAAGAUAAUGUCAAUC---- ...............((((((((((.(((((((((((((.....(((......)))....)))))).---.....)))))))...)))).))))))---- ( -25.70) >DroSec_CAF1 1899 87 + 1 ------GAACAUACCGAUUGACUUAAUGGUCGGUGCUCAAGAUAGCUUUUGAUAGCGGUGUGAGCAC---AGGGUCCGACCAAGAUAAUGUCAUUC---- ------.........((.(((((((.(((((((((((((.....(((......)))....)))))).---.....)))))))...))).)))).))---- ( -27.00) >DroSim_CAF1 3584 87 + 1 ------GAACAUACCGAUUGACUUAAUGGUCGGUGCUCAAGAUAGCUUCUGAUAGCGGUGUGAGCAC---AGGGUCCGACCAAGAUAAUGUCAAUC---- ------.........((((((((((.(((((((((((((.....(((......)))....)))))).---.....)))))))...))).)))))))---- ( -30.20) >DroEre_CAF1 1882 97 + 1 UUCCAUGAGCAUACUGAUUGACCUAAUGGUCGGUGCUCGAGAUAGCUUCUGAUAGCGGUGAGAGCAC---UGGGUCCGACCAAGAUAAGGUCAUUCCAUC ...............((.((((((..(((((((.((((......(((......)))((((....)))---)))))))))))).....)))))).)).... ( -31.00) >DroYak_CAF1 3711 93 + 1 UUCAAUAAACAUACUGAUUGACUUAAUGGUCGGUGCUCGAGAUAGCUUCUGAUAGCGGUGAGAGCAC---AGGGUCCGACCAAGAUAAAGUCAUUC---- ...............((.((((((..(((((((.((((......(((......))).((....))..---.))))))))))).....)))))).))---- ( -27.30) >DroAna_CAF1 1989 95 + 1 CCCAAUGGCGAUAUCGAUUAUCUUAAUGGAAGGUGCUCGAGAUAGCAUCUGAUAACU-UCUUAGCACGGCAGAGUUCGUUCCAGAUAAUGACGUUC---- ...((((((....((((.((((((.....)))))).))))....))(((((..(((.-.(((.((...)).)))...))).))))).....)))).---- ( -19.20) >consensus UUCAAUGAACAUACCGAUUGACUUAAUGGUCGGUGCUCAAGAUAGCUUCUGAUAGCGGUGUGAGCAC___AGGGUCCGACCAAGAUAAUGUCAUUC____ ...............((.((((....(((((((((((((.....(((......)))....)))))).........))))))).......)))).)).... (-18.40 = -18.73 + 0.34)

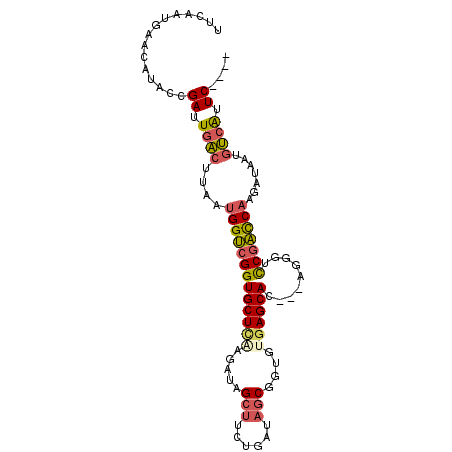
| Location | 6,854,672 – 6,854,765 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 80.48 |
| Mean single sequence MFE | -26.18 |
| Consensus MFE | -18.99 |
| Energy contribution | -19.35 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.20 |
| SVM RNA-class probability | 0.930157 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6854672 93 - 20766785 ----GAUUGACAUUAUCUUGGUCGGACCCU---GUGCUCACACCGCUAUCAGAAGCUAUCUUAAGCACCGACCAUUAACUCAAUCGGCAUGUUCAUUGAA ----((((((..(((...(((((((...((---(.((.......))...)))..(((......))).))))))).))).))))))............... ( -24.80) >DroSec_CAF1 1899 87 - 1 ----GAAUGACAUUAUCUUGGUCGGACCCU---GUGCUCACACCGCUAUCAAAAGCUAUCUUGAGCACCGACCAUUAAGUCAAUCGGUAUGUUC------ ----((.((((.(((...(((((((.....---.((((((....(((......))).....))))))))))))).))))))).)).........------ ( -26.80) >DroSim_CAF1 3584 87 - 1 ----GAUUGACAUUAUCUUGGUCGGACCCU---GUGCUCACACCGCUAUCAGAAGCUAUCUUGAGCACCGACCAUUAAGUCAAUCGGUAUGUUC------ ----(((((((.(((...(((((((.....---.((((((....(((......))).....))))))))))))).)))))))))).........------ ( -30.50) >DroEre_CAF1 1882 97 - 1 GAUGGAAUGACCUUAUCUUGGUCGGACCCA---GUGCUCUCACCGCUAUCAGAAGCUAUCUCGAGCACCGACCAUUAGGUCAAUCAGUAUGCUCAUGGAA ....((.((((((.....(((((((.....---.(((((.....(((......)))......))))))))))))..)))))).))............... ( -29.10) >DroYak_CAF1 3711 93 - 1 ----GAAUGACUUUAUCUUGGUCGGACCCU---GUGCUCUCACCGCUAUCAGAAGCUAUCUCGAGCACCGACCAUUAAGUCAAUCAGUAUGUUUAUUGAA ----...((((((.....(((((((...((---(.((.......))...)))..(((......))).)))))))..)))))).((((((....)))))). ( -29.00) >DroAna_CAF1 1989 95 - 1 ----GAACGUCAUUAUCUGGAACGAACUCUGCCGUGCUAAGA-AGUUAUCAGAUGCUAUCUCGAGCACCUUCCAUUAAGAUAAUCGAUAUCGCCAUUGGG ----....((((((((((((...(....)..))(((((.(((-(((........))).)))..))))).........))))))).)))............ ( -16.90) >consensus ____GAAUGACAUUAUCUUGGUCGGACCCU___GUGCUCACACCGCUAUCAGAAGCUAUCUCGAGCACCGACCAUUAAGUCAAUCGGUAUGUUCAUUGAA ....((.((((.(((...((((((.........((((((.....(((......)))......)))))))))))).))))))).))............... (-18.99 = -19.35 + 0.36)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:12 2006