| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,818,752 – 6,818,912 |
| Length | 160 |
| Max. P | 0.750015 |

| Location | 6,818,752 – 6,818,872 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.33 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.583126 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6818752 120 + 20766785 AAAAAAAAACCAAACUCAAAACCCACCACUAGUUGCGGUGGUCUUUGGCAAUCAAUCGUGGCUUGGCCAAAUACUAGACGAGUAUUUAGGGCCAGCGAAGCGAAGAUCACAAGAAGCUGA ................((((..(((((.(.....).)))))..))))((......((((.(((.((((.((((((.....))))))...)))))))...))))...((....)).))... ( -32.80) >DroSec_CAF1 20687 119 + 1 CCAG-AAAACCAAACUUAAAACCCACCACUAGUUGCGGUGGUCUUUGGCAAUCAAUCGUGGUUUGGCCAAAUACUAGACGAGUAGUUAGGGCCAGCAAAGCGAAGAUCACAAGAAGCUGA .(((-.................(((((.(.....).)))))((((.(...(((..((((.(((.((((...((((.....)))).....)))))))...)))).)))..)))))..))). ( -29.80) >DroSim_CAF1 21226 119 + 1 CCAG-AAAACCAAACUCAAAACCCACCACUAGUUGCGGUGGUCUUUGGCAAUCAAUCGUGGUUUGGCCAAAUACUAGACGAGUAUUUAGGGCCAGCAAAGCGAAGAUCACAAGAAGCUGA .(((-..........(((((..(((((.(.....).)))))..))))).........(((((((((((.((((((.....))))))...)))).((...))..)))))))......))). ( -33.40) >DroEre_CAF1 20430 119 + 1 CCGG-AAAACCAAACUCAAAACCCACCACUAGUUGGGGCGGUCUUUGGCAAUUAAUGGUGGUUUGGCCAAAUACUAGACGAGUAUUUAGGGUCAGCAAGGCGAAGAUCACAUAAAGCUGA .(((-................((((.(....).))))(.((((((((.((((((....)))))(((((.((((((.....))))))...)))))....).)))))))).)......))). ( -28.00) >consensus CCAG_AAAACCAAACUCAAAACCCACCACUAGUUGCGGUGGUCUUUGGCAAUCAAUCGUGGUUUGGCCAAAUACUAGACGAGUAUUUAGGGCCAGCAAAGCGAAGAUCACAAGAAGCUGA .............................(((((...((((((((((((........)).(((.((((.((((((.....))))))...)))))))....))))))))))....))))). (-28.20 = -28.33 + 0.12)


| Location | 6,818,752 – 6,818,872 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.89 |
| Mean single sequence MFE | -33.90 |
| Consensus MFE | -30.91 |
| Energy contribution | -31.10 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.750015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6818752 120 - 20766785 UCAGCUUCUUGUGAUCUUCGCUUCGCUGGCCCUAAAUACUCGUCUAGUAUUUGGCCAAGCCACGAUUGAUUGCCAAAGACCACCGCAACUAGUGGUGGGUUUUGAGUUUGGUUUUUUUUU ..((((.((.(..(((.(((....(((((((..(((((((.....))))))))))).)))..)))..)))..)(((((.(((((((.....))))))).)))))))...))))....... ( -37.50) >DroSec_CAF1 20687 119 - 1 UCAGCUUCUUGUGAUCUUCGCUUUGCUGGCCCUAACUACUCGUCUAGUAUUUGGCCAAACCACGAUUGAUUGCCAAAGACCACCGCAACUAGUGGUGGGUUUUAAGUUUGGUUUU-CUGG .(((..(((((..(((.(((......(((((...((((......))))....))))).....)))..)))..)..))))(((((((.....))))))).................-))). ( -31.80) >DroSim_CAF1 21226 119 - 1 UCAGCUUCUUGUGAUCUUCGCUUUGCUGGCCCUAAAUACUCGUCUAGUAUUUGGCCAAACCACGAUUGAUUGCCAAAGACCACCGCAACUAGUGGUGGGUUUUGAGUUUGGUUUU-CUGG .(((......(..(((.(((......(((((..(((((((.....)))))))))))).....)))..)))..)(((((.(((((((.....))))))).)))))...........-))). ( -35.90) >DroEre_CAF1 20430 119 - 1 UCAGCUUUAUGUGAUCUUCGCCUUGCUGACCCUAAAUACUCGUCUAGUAUUUGGCCAAACCACCAUUAAUUGCCAAAGACCGCCCCAACUAGUGGUGGGUUUUGAGUUUGGUUUU-CCGG (((((.....(((.....)))...)))))(((((((((((.....)))))))))((((((...((.....)).(((((.(((((.(.....).))))).))))).))))))....-..)) ( -30.40) >consensus UCAGCUUCUUGUGAUCUUCGCUUUGCUGGCCCUAAAUACUCGUCUAGUAUUUGGCCAAACCACGAUUGAUUGCCAAAGACCACCGCAACUAGUGGUGGGUUUUGAGUUUGGUUUU_CUGG (((((.....(((.....)))...)))))..(((((((((.....)))))))))((((((((........)).(((((.(((((((.....))))))).))))).))))))......... (-30.91 = -31.10 + 0.19)

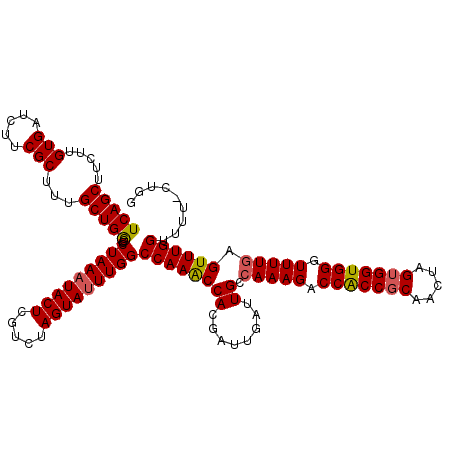
| Location | 6,818,792 – 6,818,912 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -34.00 |
| Consensus MFE | -32.14 |
| Energy contribution | -32.33 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.95 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6818792 120 + 20766785 GUCUUUGGCAAUCAAUCGUGGCUUGGCCAAAUACUAGACGAGUAUUUAGGGCCAGCGAAGCGAAGAUCACAAGAAGCUGACUGGGCGACAUAGCUGCAAUUGAGUGAGUAAAAUGACUGA ((((((..((.(((((.(..((((((((.((((((.....))))))...)))))..(..((..((.(((........)))))..))..)..)))..).))))).))....))).)))... ( -32.70) >DroSec_CAF1 20726 120 + 1 GUCUUUGGCAAUCAAUCGUGGUUUGGCCAAAUACUAGACGAGUAGUUAGGGCCAGCAAAGCGAAGAUCACAAGAAGCUGACUGGGCGACAUAGCUGCGAUUGAGUGAGCAAAAUGACUGA (((((((.((.(((((((..((((((((...((((.....)))).....))))).....((..((.(((........)))))..)).....)))..))))))).))..))))..)))... ( -35.60) >DroSim_CAF1 21265 120 + 1 GUCUUUGGCAAUCAAUCGUGGUUUGGCCAAAUACUAGACGAGUAUUUAGGGCCAGCAAAGCGAAGAUCACAAGAAGCUGACUGGGCGACAUAGCUGCGAUUGAGUGAGUAAAAUGACUGA ((((((..((.(((((((..((((((((.((((((.....))))))...))))).....((..((.(((........)))))..)).....)))..))))))).))....))).)))... ( -35.90) >DroEre_CAF1 20469 120 + 1 GUCUUUGGCAAUUAAUGGUGGUUUGGCCAAAUACUAGACGAGUAUUUAGGGUCAGCAAGGCGAAGAUCACAUAAAGCUGACUGGGCGACAUAGCUGCGAUUGAGUGAAUAAAAUGACUGA ((((((..((.(((((.(..((((.((((((((((.....)))))))..(((((((...(.(.....).).....))))))).))).)...)))..).))))).))....))).)))... ( -31.80) >consensus GUCUUUGGCAAUCAAUCGUGGUUUGGCCAAAUACUAGACGAGUAUUUAGGGCCAGCAAAGCGAAGAUCACAAGAAGCUGACUGGGCGACAUAGCUGCGAUUGAGUGAGUAAAAUGACUGA ((((((..((.(((((((..((((((((.((((((.....))))))...))))).....((..((.(((........)))))..)).....)))..))))))).))....))).)))... (-32.14 = -32.33 + 0.19)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:14:01 2006