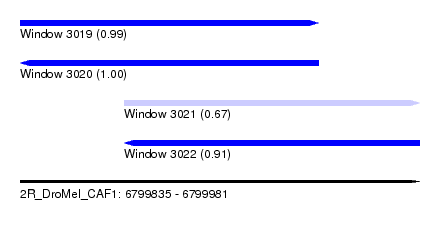
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,799,835 – 6,799,981 |
| Length | 146 |
| Max. P | 0.996506 |

| Location | 6,799,835 – 6,799,944 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -29.05 |
| Consensus MFE | -27.30 |
| Energy contribution | -27.50 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.94 |
| SVM decision value | 2.52 |
| SVM RNA-class probability | 0.994911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6799835 109 + 20766785 UCUCUCCCCAUUCGGUCCUCCCACCUCUCAGGCACUCCUCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUGUGCACACCGUGCCGGCUGUUCAACAGAGU .((((........(((......)))...((((..................)))).((((((((((((.((((......)).)))))))).))))))........)))). ( -30.17) >DroSec_CAF1 5915 109 + 1 UCUCUCCCCAUUCGGUCCUCCCACCUCUCAGGCACUCCUCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUUGCACACCGUGCCGGCUGUUCAACAGAGU .((((........(((......)))...((((..................)))).((((((((((((.(((....))).....)))))).))))))........)))). ( -29.37) >DroSim_CAF1 5909 109 + 1 UCUCUCCCCAUUCGGUCCUCCCACCUCUCAGGCACUCCUCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUUGCACACCGUGCCGGCUGUUCAACAGAGU .((((........(((......)))...((((..................)))).((((((((((((.(((....))).....)))))).))))))........)))). ( -29.37) >DroEre_CAF1 5915 108 + 1 UCUAUCCCCAUUCGGUCCUCCCACCUCUCUGUCACUCCACCACUCCUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUU-GCACACCGUGCCGGCUGUUCAACAAAGU .............(((......)))..............................((((((((((((.(((....)))..-..)))))).))))))............. ( -26.50) >DroYak_CAF1 5921 109 + 1 UCUCUCCCCAUUCGGUCCUCCCACCUCUCUGUCACUCCUCCACUCCUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUCGCACACCGUGCCGGCUGUUCAACAGAGU .............(((......))).((((((.....................((((((((((((((.(((....))).....)))))).)))))).))...)))))). ( -29.85) >consensus UCUCUCCCCAUUCGGUCCUCCCACCUCUCAGGCACUCCUCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUUGCACACCGUGCCGGCUGUUCAACAGAGU .((((........(((......)))..............................((((((((((((.(((....))).....)))))).))))))........)))). (-27.30 = -27.50 + 0.20)


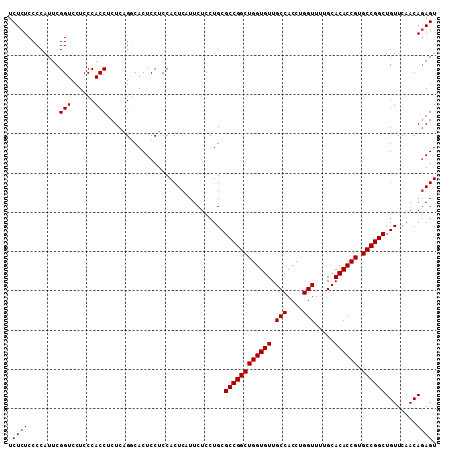
| Location | 6,799,835 – 6,799,944 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 96.24 |
| Mean single sequence MFE | -38.12 |
| Consensus MFE | -34.54 |
| Energy contribution | -34.38 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.91 |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.996506 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6799835 109 - 20766785 ACUCUGUUGAACAGCCGGCACGGUGUGCACAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGAGGAGUGCCUGAGAGGUGGGAGGACCGAAUGGGGAGAGA .(((..((.....((((((..(((.......))).((((....)))).)))))).((......))))..))).....(((.(..(((......)))...).)))..... ( -39.00) >DroSec_CAF1 5915 109 - 1 ACUCUGUUGAACAGCCGGCACGGUGUGCAAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGAGGAGUGCCUGAGAGGUGGGAGGACCGAAUGGGGAGAGA .(((..((.....((((((..(((.......))).((((....)))).)))))).((......))))..))).....(((.(..(((......)))...).)))..... ( -39.00) >DroSim_CAF1 5909 109 - 1 ACUCUGUUGAACAGCCGGCACGGUGUGCAAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGAGGAGUGCCUGAGAGGUGGGAGGACCGAAUGGGGAGAGA .(((..((.....((((((..(((.......))).((((....)))).)))))).((......))))..))).....(((.(..(((......)))...).)))..... ( -39.00) >DroEre_CAF1 5915 108 - 1 ACUUUGUUGAACAGCCGGCACGGUGUGC-AAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAGGAGUGGUGGAGUGACAGAGAGGUGGGAGGACCGAAUGGGGAUAGA .((((((((..((((((((..(((....-..))).((((....)))).))))))((..........))..))...)))))))).(((......)))............. ( -36.10) >DroYak_CAF1 5921 109 - 1 ACUCUGUUGAACAGCCGGCACGGUGUGCGAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAGGAGUGGAGGAGUGACAGAGAGGUGGGAGGACCGAAUGGGGAGAGA .((((((((..(.((((((..(((.......))).((((....)))).))))))((..........))...)...)))))))).(((......)))............. ( -37.50) >consensus ACUCUGUUGAACAGCCGGCACGGUGUGCAAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGAGGAGUGCCUGAGAGGUGGGAGGACCGAAUGGGGAGAGA .(((..((.....((((((..(((.......))).((((....)))).))))))..............................(((......))).))..)))..... (-34.54 = -34.38 + -0.16)

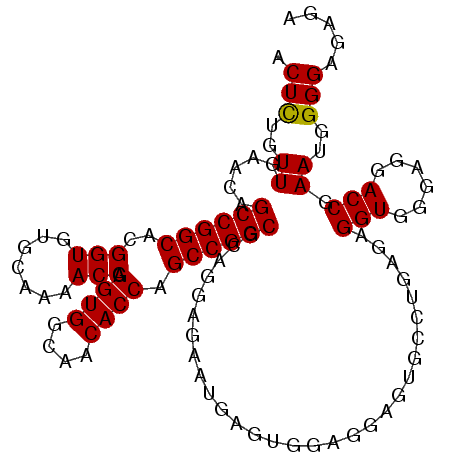
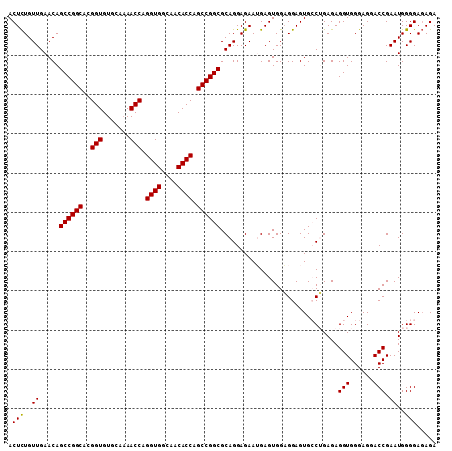
| Location | 6,799,873 – 6,799,981 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -33.35 |
| Consensus MFE | -25.71 |
| Energy contribution | -25.27 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.668706 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6799873 108 + 20766785 UCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUGUGCACACCGUGCCGGCUGUUCAACAGAGUUGACACUUCUUGUUCAGAAAGGUGACUCGUUC---CGGAA (((..............((((((((((((.((((......)).)))))))).)))))).........(((((.((..((((.....))))..)))))))....---.))). ( -36.60) >DroSec_CAF1 5953 108 + 1 UCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUUGCACACCGUGCCGGCUGUUCAACAGAGUUGACACUUCUUGUUCAAAAAGGUGACUCGUUC---CGGAA (((..............((((((((((((.(((....))).....)))))).)))))).........((((...(((((.((.....)).)))))))))....---.))). ( -34.40) >DroSim_CAF1 5947 108 + 1 UCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUUGCACACCGUGCCGGCUGUUCAACAGAGUUGACACUUCUUGUUCAAAAAGGUGACUCGUUC---CGGAA (((..............((((((((((((.(((....))).....)))))).)))))).........((((...(((((.((.....)).)))))))))....---.))). ( -34.40) >DroEre_CAF1 5953 107 + 1 ACCACUCCUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUU-GCACACCGUGCCGGCUGUUCAACAAAGUUGACACUUCUUGUUCGGGACGGUGACUUGUUC---CGGAA .................((((((((((((.(((....)))..-..)))))).))))))(((.((((...))))))).......(((((((((....)..))))---)))). ( -38.30) >DroYak_CAF1 5959 91 + 1 UCCACUCCUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUCGCACACCGUGCCGGCUGUUCAACAGAGUUGACACUUCUUG-----------------UUC---CGGAA (((..............((((((((((((.(((....))).....)))))).))))))(((.((((...))))))).......-----------------...---.))). ( -30.00) >DroAna_CAF1 15352 104 + 1 -----UCUUUCUCUUUCUCCGGUCGGUGUUGCCACCUGGGGACGGAAGAUGUGCCGGCUGUUGAAUAAAGUUUACAAUA--AGCUCCGAAAAUGAACUAGCUCAUUGGGAA -----...(((((.(((..(((((((..(......(((....))).....)..)))))))..)))...((((((...))--)))).....(((((......)))))))))) ( -26.40) >consensus UCCACUCAUUCUCCUGCGCCGGCUGGUGUUGCCACCUGGUUUUGCACACCGUGCCGGCUGUUCAACAGAGUUGACACUUCUUGUUCAGAAAGGUGACUCGUUC___CGGAA ...........(((...((((((((((((.(((....))).....)))))).))))))(((.((((...)))))))...............................))). (-25.71 = -25.27 + -0.44)
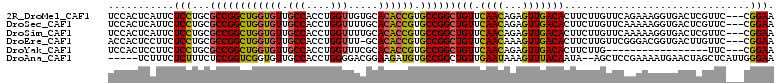
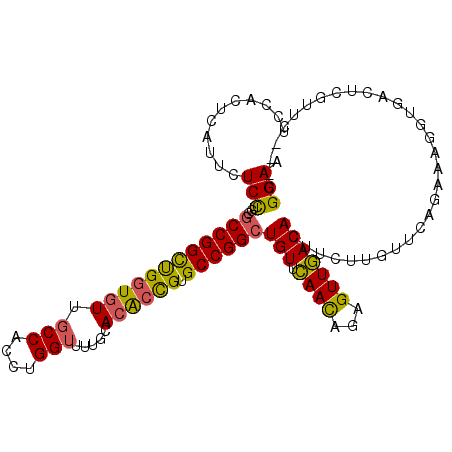

| Location | 6,799,873 – 6,799,981 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -36.48 |
| Consensus MFE | -26.62 |
| Energy contribution | -27.95 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.09 |
| SVM RNA-class probability | 0.913695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6799873 108 - 20766785 UUCCG---GAACGAGUCACCUUUCUGAACAAGAAGUGUCAACUCUGUUGAACAGCCGGCACGGUGUGCACAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGA .....---...(.(.(((..((((((........((.(((((...))))))).((((((..(((.......))).((((....)))).)))))).))))))..))).).). ( -39.30) >DroSec_CAF1 5953 108 - 1 UUCCG---GAACGAGUCACCUUUUUGAACAAGAAGUGUCAACUCUGUUGAACAGCCGGCACGGUGUGCAAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGA ((((.---.(((((((.((((((((....)))))).))..)))).))).....((((((..(((.......))).((((....)))).))))))...)))).......... ( -39.10) >DroSim_CAF1 5947 108 - 1 UUCCG---GAACGAGUCACCUUUUUGAACAAGAAGUGUCAACUCUGUUGAACAGCCGGCACGGUGUGCAAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGA ((((.---.(((((((.((((((((....)))))).))..)))).))).....((((((..(((.......))).((((....)))).))))))...)))).......... ( -39.10) >DroEre_CAF1 5953 107 - 1 UUCCG---GAACAAGUCACCGUCCCGAACAAGAAGUGUCAACUUUGUUGAACAGCCGGCACGGUGUGC-AAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAGGAGUGGU .....---.........((((..((...((((((((....))))).)))....((((((..(((....-..))).((((....)))).)))))).........))..)))) ( -38.90) >DroYak_CAF1 5959 91 - 1 UUCCG---GAA-----------------CAAGAAGUGUCAACUCUGUUGAACAGCCGGCACGGUGUGCGAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAGGAGUGGA (((((---..(-----------------(.....))..)...(((.(((....((((((..(((.......))).((((....)))).)))))).))).))).)))).... ( -35.50) >DroAna_CAF1 15352 104 - 1 UUCCCAAUGAGCUAGUUCAUUUUCGGAGCU--UAUUGUAAACUUUAUUCAACAGCCGGCACAUCUUCCGUCCCCAGGUGGCAACACCGACCGGAGAAAGAGAAAGA----- ....(((((((((.(........)..))))--)))))....((((.(((....(....)...(((((.(((....((((....))))))).)))))..))))))).----- ( -27.00) >consensus UUCCG___GAACGAGUCACCUUUCUGAACAAGAAGUGUCAACUCUGUUGAACAGCCGGCACGGUGUGCAAAACCAGGUGGCAACACCAGCCGGCGCAGGAGAAUGAGUGGA ((((...............................(((((((...)))).)))((((((..(((.......))).((((....)))).))))))...)))).......... (-26.62 = -27.95 + 1.33)

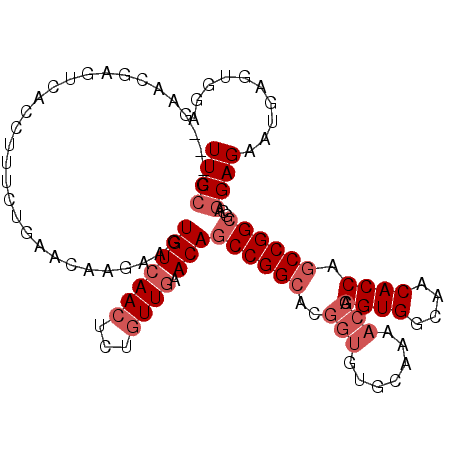
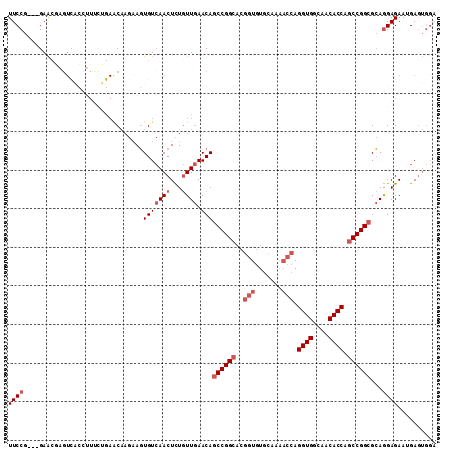
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:57 2006