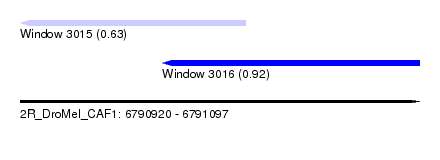
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,790,920 – 6,791,097 |
| Length | 177 |
| Max. P | 0.917753 |

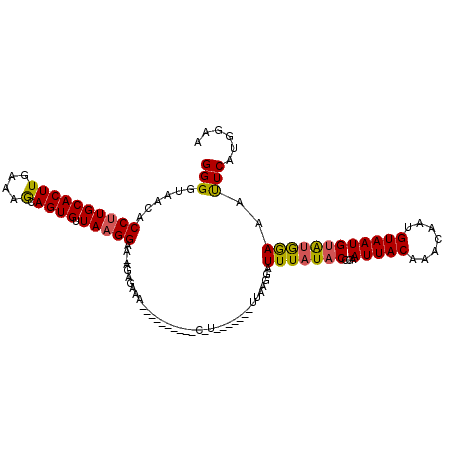
| Location | 6,790,920 – 6,791,020 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 88.36 |
| Mean single sequence MFE | -26.03 |
| Consensus MFE | -19.65 |
| Energy contribution | -20.15 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.631340 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6790920 100 - 20766785 GAAUUACAAACGAAGUAAUGUAUGGAAAUUCAUGGAA------------CUUUCUUCUCAGGACGAAGACCUGACCCCCGAGCAGAUUGCCGUUCUGCAGAAGGCAUUCAAC ((((.......(((((.(((.((....)).)))...)------------))))((((((((((((..((.(((.(......)))).))..))))))).)))))..))))... ( -23.20) >DroSec_CAF1 41032 100 - 1 GAAUUACAAGCAAAGUAAUGUAUGGAAAUUCAUGGAA------------CUUUCUCCUCAGGACGAAGAUCUGACCCCCGAGCAGAUUGCCGUUCUGCAGAAGGCAUUCAAC (((((.((.(((......))).))..))))).((((.------------((((((...(((((((..((((((.(......)))))))..))))))).))))))..)))).. ( -27.40) >DroSim_CAF1 49974 100 - 1 GAAUUACAAACAAUGUAAUGUAUGGAAAUUCAUGGAA------------CUUUCUCCUCAGGACGAAGAUCUGACCCCCGAGCAGAUUGCCGUUCUGCAGAAGGCAUUCAAC (((((.((.(((......))).))..))))).((((.------------((((((...(((((((..((((((.(......)))))))..))))))).))))))..)))).. ( -27.10) >DroYak_CAF1 48921 112 - 1 AAAUUACAAACAAUGUAAUGUGUAAAAACUCACUGAACGCAUUUCUAUGCUUUCUCCUCAGGACGAAGACCUGACCCCCGAGCAGAUUGCCGUUCUGCAGAAGGCAUUCAAC ..((((((.....))))))((((.............))))......(((((((((..(((((.......))))).......(((((.......))))))))))))))..... ( -26.42) >consensus GAAUUACAAACAAAGUAAUGUAUGGAAAUUCAUGGAA____________CUUUCUCCUCAGGACGAAGACCUGACCCCCGAGCAGAUUGCCGUUCUGCAGAAGGCAUUCAAC ..(((((.......)))))..............................((((((...(((((((..((((((.(......)))))))..))))))).))))))........ (-19.65 = -20.15 + 0.50)

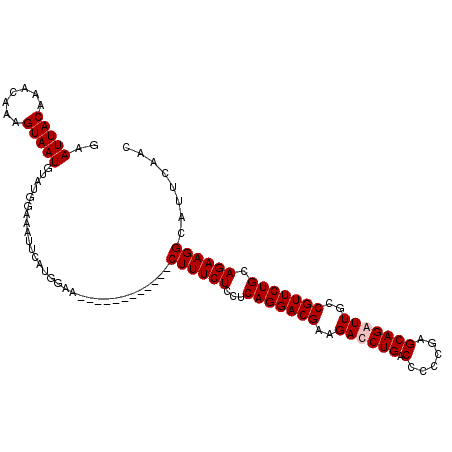
| Location | 6,790,983 – 6,791,097 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.04 |
| Mean single sequence MFE | -22.70 |
| Consensus MFE | -11.73 |
| Energy contribution | -11.63 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.917753 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6790983 114 - 20766785 GGGGUAACACCUUGCACUUGAAAGCAGUGUUAAGGAAUAGAGAAAUGUCGUUUCUCAUCA---AUUUAAAGAUUUUUACCGAAUUACAAACGAAGUAAUGUAUGGAAAUUCAUGGAA ..(((((..((((((((((....).)))).)))))....(((((((...)))))))....---............)))))(((((.((.(((......))).))..)))))...... ( -23.40) >DroSim_CAF1 50037 88 - 1 GGGGUAACUCCUUGCACUUGAAAGCAGUGUUAAGGA-----------------------------UUAAGGAUUUAUUCUGAAUUACAAACAAUGUAAUGUAUGGAAAUUCAUGGAA (((((((.((((((..(((((........)))))..-----------------------------.)))))).)))))))(((((.((.(((......))).))..)))))...... ( -18.80) >DroYak_CAF1 48996 109 - 1 GGGCAAACACCUUGCACUUGAAAA-AGUGUUAUGGAACAGAGAAAAUC-------CGUUUUUUUAUCACGGAUUUAUACCAAAUUACAAACAAUGUAAUGUGUAAAAACUCACUGAA .........((..((((((....)-)))))...))..(((.(((((((-------(((.........)))))))).(((...((((((.....))))))..))).....)).))).. ( -25.90) >consensus GGGGUAACACCUUGCACUUGAAAGCAGUGUUAAGGAA_AGAGAAA__________C_U_______UUAAGGAUUUAUACCGAAUUACAAACAAUGUAAUGUAUGGAAAUUCAUGGAA (((......((((((((((....).)))).))))).....................................(((((((...(((((.......))))))))))))..)))...... (-11.73 = -11.63 + -0.10)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:51 2006