| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,790,325 – 6,790,431 |
| Length | 106 |
| Max. P | 0.975201 |

| Location | 6,790,325 – 6,790,431 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 69.75 |
| Mean single sequence MFE | -34.75 |
| Consensus MFE | -15.99 |
| Energy contribution | -15.63 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.46 |
| SVM decision value | 0.70 |
| SVM RNA-class probability | 0.827379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6790325 106 + 20766785 ---------UCUCCCCUCG-UUGGUGGGAUGGUGGUGUUAGUGGGGAGGGGUAGG--GGUUCUCCACCG-CAACAAAUUGAUAUAAUUUGUGUGGAAA-AUACCAUGGCAGCGCGCCAAG ---------((((.((...-..)).))))(((((((....((((((((((.....--..)))))).(((-((.(((((((...))))))))))))...-...))))....)).))))).. ( -35.60) >DroSec_CAF1 40444 93 + 1 ---------UCUCCCCUCG-UCGGUGGGAAUG----------GGGGAGGGCGGU---GG--CUCCAUCG-CAACAAAUUGAUAUAAUUUGUGUGGAAA-AUACCAAGGCAGCGGGCCAAG ---------((((((((..-((.....))..)----------)))))))(((((---(.--...)))))-).((((((((...)))))))).(((...-...))).(((.....)))... ( -33.70) >DroSim_CAF1 49391 93 + 1 ---------UCUCCCCUCG-UAGGUGGGAAUG----------GGGGAGGGCGAU---GG--CUCCACCG-CAACAAAUUGAUAUAAUUUGUGUGGAAA-AUACCAUGGCAGCGGGCCAAG ---------.((((((.((-(........)))----------))))))(((..(---(.--...))(((-(.((((((((...))))))))((((...-...))))....)))))))... ( -32.70) >DroEre_CAF1 48169 101 + 1 ---------UCUCCCCUCU-UCAGCGGGAUUAGGAUG-----GGGGAGGGGGAGGGGGG--CUCCGCCG-CAACAAAUUGAUAUAAUUUGUGUGGAAA-AUACCAUGGCAGCGGGCCAAG ---------((((((((((-((..(........)...-----))))).)))))))..((--(.((((.(-(.((((((((...))))))))((((...-...)))).)).)))))))... ( -40.30) >DroYak_CAF1 48339 89 + 1 ---------UAUCCCCUCG-UCAGCGGGUU-----------------GGGGGCGGGGGG--CUCCACCG-CAACAAAUUGAUAUAAUUUGUGUGGAAA-AUACCUUGGCAGGGGGCCAAG ---------..((((((.(-((((.((...-----------------.(((((.....)--)))).(((-((.(((((((...))))))))))))...-...)))))))))))))..... ( -35.60) >DroPer_CAF1 38295 104 + 1 GAACGACAUUGUCUUCUCGGCCAGCCUGAAC-------------GGAGAACAGC---GGGCCUGUAGAGCCUACAAAUUGAUAUAAUUUGCUUCUUCCUCAACGCUGGCACGGGGCCAAA ....(((...))).(((((((((((.(((..-------------((((((....---(((((....).)))).(((((((...))))))).)))))).)))..)))))).)))))..... ( -30.60) >consensus _________UCUCCCCUCG_UCAGCGGGAAUG__________GGGGAGGGCGAG___GG__CUCCACCG_CAACAAAUUGAUAUAAUUUGUGUGGAAA_AUACCAUGGCAGCGGGCCAAG .............(((((...........................)))))............(((((.....((((((((...))))))))))))).........((((.....)))).. (-15.99 = -15.63 + -0.36)

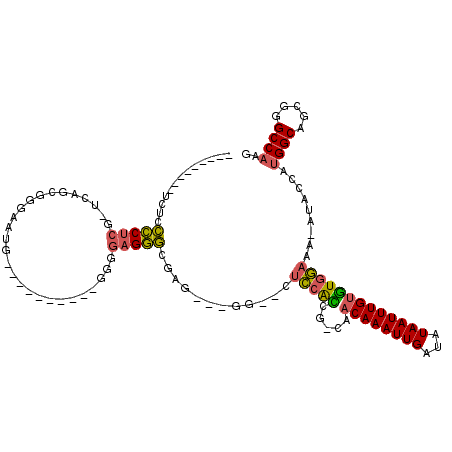
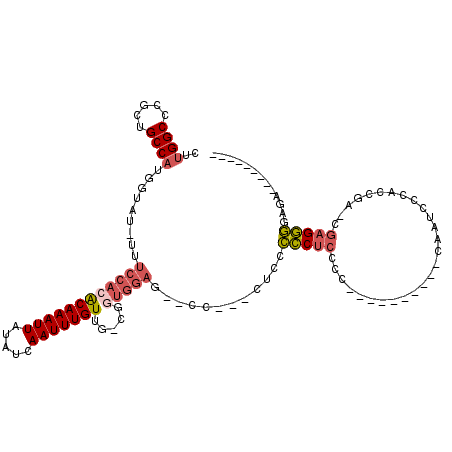
| Location | 6,790,325 – 6,790,431 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 69.75 |
| Mean single sequence MFE | -30.28 |
| Consensus MFE | -13.27 |
| Energy contribution | -14.08 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.44 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975201 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6790325 106 - 20766785 CUUGGCGCGCUGCCAUGGUAU-UUUCCACACAAAUUAUAUCAAUUUGUUG-CGGUGGAGAACC--CCUACCCCUCCCCACUAACACCACCAUCCCACCAA-CGAGGGGAGA--------- ..((((.....)))).((..(-((((((((((((((.....)))))))..-..))))))))..--)).....((((((......................-...)))))).--------- ( -29.81) >DroSec_CAF1 40444 93 - 1 CUUGGCCCGCUGCCUUGGUAU-UUUCCACACAAAUUAUAUCAAUUUGUUG-CGAUGGAG--CC---ACCGCCCUCCCC----------CAUUCCCACCGA-CGAGGGGAGA--------- ..(((((((.(((..(((...-...))).(((((((.....))))))).)-)).))).)--))---).....((((((----------...((.....))-...)))))).--------- ( -27.70) >DroSim_CAF1 49391 93 - 1 CUUGGCCCGCUGCCAUGGUAU-UUUCCACACAAAUUAUAUCAAUUUGUUG-CGGUGGAG--CC---AUCGCCCUCCCC----------CAUUCCCACCUA-CGAGGGGAGA--------- ..(((((((((((..(((...-...))).(((((((.....))))))).)-)))))).)--))---).....((((((----------(...........-.).)))))).--------- ( -33.50) >DroEre_CAF1 48169 101 - 1 CUUGGCCCGCUGCCAUGGUAU-UUUCCACACAAAUUAUAUCAAUUUGUUG-CGGCGGAG--CCCCCCUCCCCCUCCCC-----CAUCCUAAUCCCGCUGA-AGAGGGGAGA--------- ...((((((((((..(((...-...))).(((((((.....))))))).)-)))))).)--)).........((((((-----(.((...........))-.).)))))).--------- ( -34.50) >DroYak_CAF1 48339 89 - 1 CUUGGCCCCCUGCCAAGGUAU-UUUCCACACAAAUUAUAUCAAUUUGUUG-CGGUGGAG--CCCCCCGCCCCC-----------------AACCCGCUGA-CGAGGGGAUA--------- ((((((.....))))))....-...............((((..(((((((-((((((.(--(.....))..))-----------------)..)))).))-))))..))))--------- ( -25.70) >DroPer_CAF1 38295 104 - 1 UUUGGCCCCGUGCCAGCGUUGAGGAAGAAGCAAAUUAUAUCAAUUUGUAGGCUCUACAGGCCC---GCUGUUCUCC-------------GUUCAGGCUGGCCGAGAAGACAAUGUCGUUC (((((((....(((((((..(((((...(((....((((......))))((((.....)))).---))).))))))-------------)))..))).)))))))..(((...))).... ( -30.50) >consensus CUUGGCCCGCUGCCAUGGUAU_UUUCCACACAAAUUAUAUCAAUUUGUUG_CGGUGGAG__CC___CUCCCCCUCCCC__________CAAUCCCACCGA_CGAGGGGAGA_________ ..((((.....)))).........((((((((((((.....))))))).....)))))............(((((...........................)))))............. (-13.27 = -14.08 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:48 2006