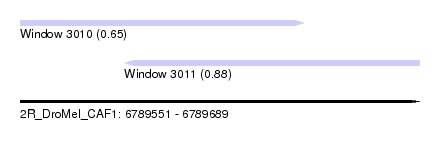
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,789,551 – 6,789,689 |
| Length | 138 |
| Max. P | 0.884052 |

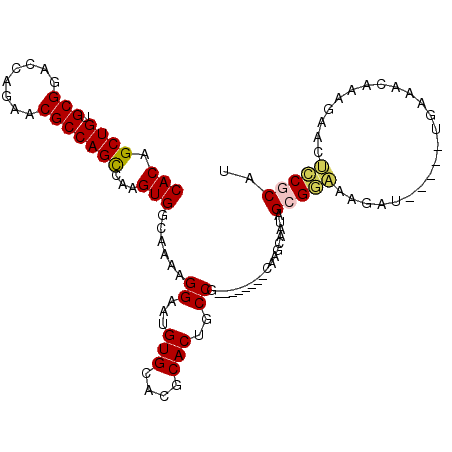
| Location | 6,789,551 – 6,789,649 |
|---|---|
| Length | 98 |
| Sequences | 5 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 82.72 |
| Mean single sequence MFE | -30.96 |
| Consensus MFE | -20.48 |
| Energy contribution | -20.64 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.650940 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
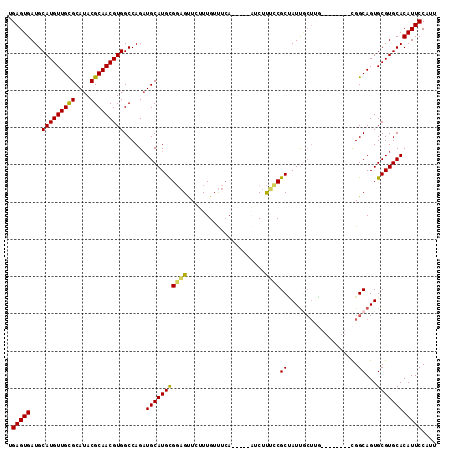
>2R_DroMel_CAF1 6789551 98 + 20766785 CACAGCUGUGCGGACCAGAACGCCAGUCAAGUGGCAAACGGAAUGUGCACGCACUGCCG--------CAAGUAAUAGCGGAAAGAU-----UGAAACAAAGAACUCCGCAU ....((((.(((........)))))))...((((((........(((....))))))))--------)........(((((....(-----(......))....))))).. ( -26.90) >DroSec_CAF1 39699 98 + 1 CACAGCUGUGCGGACCAGAACGCCAGCCAAGUGGCAAAUGGAAUGUGCACGCACUGCCG--------CAAGCAAUGGCGGAAAGAU-----UGAAACAAAGAACUCCGCAU ....(((.(((((.(((....((((......))))...)))...(((....)))..)))--------)))))....(((((....(-----(......))....))))).. ( -30.50) >DroSim_CAF1 48613 98 + 1 CACAGCUGUGCGGACCAGAACGCCAGCCAAGUGGCAAAUGGAAUGUGCACGCACUGCCG--------CAAGCAAUAGCGGAAAGAU-----UGAAACAAAGAACUCCGCAU ....(((.(((((.(((....((((......))))...)))...(((....)))..)))--------)))))....(((((....(-----(......))....))))).. ( -30.70) >DroEre_CAF1 42264 100 + 1 CACAGCUGUGCGGACCAGAACGCCAGCCAAGUGGCAAAAGGAAUGUGCACGCACUGCCGGC-----------AAUUGCGCCAAGAACGAACUCAGACAAUCAGCGCCACAU ....((((.(((........)))))))...(((((....((...(((....)))..))(((-----------......))).......................))))).. ( -31.60) >DroYak_CAF1 47464 111 + 1 CACAGCUGUGCGGACCAGAACGCCAGCCAAGUGGCAAAAGGAAUGUGCACGCACUGCCAGCGAUCGGGCAGCGAUCGCGCCGAGAAUGUACUGAAACAAAGAACUGCACAU .(((((((.(((........)))))))....((((....((...(((....)))..)).(((((((.....)))))))))))....)))...................... ( -35.10) >consensus CACAGCUGUGCGGACCAGAACGCCAGCCAAGUGGCAAAAGGAAUGUGCACGCACUGCCG________CAAGCAAUAGCGGAAAGAU_____UGAAACAAAGAACUCCGCAU (((.((((.(((........)))))))...)))......((...(((....)))..))..................(((((.......................))))).. (-20.48 = -20.64 + 0.16)


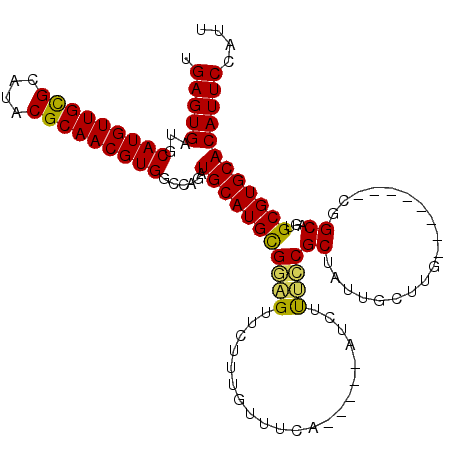
| Location | 6,789,587 – 6,789,689 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 82.98 |
| Mean single sequence MFE | -37.39 |
| Consensus MFE | -28.19 |
| Energy contribution | -27.55 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884052 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6789587 102 - 20766785 UGAGUGAUGCAUGUUGCGCAUACGCAACGUGGCCAGAUGCAUGCGGAGUUCUUUGUUUCA-----AUCUUUCCGCUAUUACUUG--------CGGCAGUGCGUGCACAUUCCGUU .(((((...(((((((((....)))))))))......(((((((((((........))).-----......((((........)--------)))...))))))))))))).... ( -34.80) >DroSec_CAF1 39735 102 - 1 UGAGUGAUGCAUGUUGUGCAUACGCAACGUGGCCAGAUGCAUGCGGAGUUCUUUGUUUCA-----AUCUUUCCGCCAUUGCUUG--------CGGCAGUGCGUGCACAUUCCAUU .(((((.(((((((..(((...(((((((((((.((((....(((((....)))))....-----))))....)))).)).)))--------)))))..)))))))))))).... ( -34.70) >DroSim_CAF1 48649 102 - 1 UGAGUGAUGCAUGUUGCGCAUACGCAACGUGGCCAGAUGCAUGCGGAGUUCUUUGUUUCA-----AUCUUUCCGCUAUUGCUUG--------CGGCAGUGCGUGCACAUUCCAUU .(((((.(((((((((((....))))))((.(((....(((.((((((....(((...))-----)...))))))...)))...--------.)))...)))))))))))).... ( -36.30) >DroEre_CAF1 42300 104 - 1 UGAGUGAUGCAUGUUGCGCAUACGCAACGUGGCCAGUUGCAUGUGGCGCUGAUUGUCUGAGUUCGUUCUUGGCGCAAUU-----------GCCGGCAGUGCGUGCACAUUCCUUU .(((((.(((((((.(((((((.(((((.......))))).))).))))..((((((.(((......)))((((....)-----------))))))))))))))))))))).... ( -38.60) >DroYak_CAF1 47500 115 - 1 UGAGUGAUGCAUGUUGCGCAUACGCAACGUGGCCAGAUGCAUGUGCAGUUCUUUGUUUCAGUACAUUCUCGGCGCGAUCGCUGCCCGAUCGCUGGCAGUGCGUGCACAUUCCUUU .(((((...(((((((((....)))))))))((((((...((((((..............))))))))).)))(((..(((((((.(....).)))))))..))).))))).... ( -42.54) >consensus UGAGUGAUGCAUGUUGCGCAUACGCAACGUGGCCAGAUGCAUGCGGAGUUCUUUGUUUCA_____AUCUUUCCGCUAUUGCUUG________CGGCAGUGCGUGCACAUUCCAUU .(((((...(((((((((....)))))))))......(((((((((((.....................))))((...................))...)))))))))))).... (-28.19 = -27.55 + -0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:47 2006