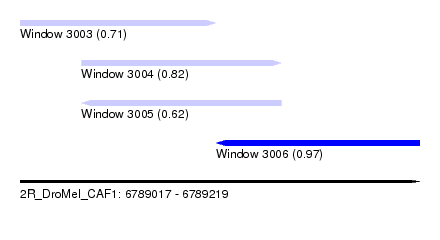
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,789,017 – 6,789,219 |
| Length | 202 |
| Max. P | 0.972047 |

| Location | 6,789,017 – 6,789,116 |
|---|---|
| Length | 99 |
| Sequences | 5 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 86.16 |
| Mean single sequence MFE | -28.74 |
| Consensus MFE | -25.16 |
| Energy contribution | -25.52 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705204 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6789017 99 + 20766785 CGGUGUCAAGAAUCGCAUAUUGAGAAGUCUGCCCAGGCGGAUGUCAAUAAAUCUGACACCCAACACAAUCCGCCACAUUUUCAGCUCUUUCACAGGGCG .(((((((.((......((((((...((((((....)))))).))))))..))))))))).......................(((((.....))))). ( -28.70) >DroSec_CAF1 39165 99 + 1 CGGUGUCAAGAAUCGCAUGUUGAGAAGUCUGCCCAGGCGGAUGUCAAUAAAUCUGACACCCAACGGAAUCCGCCGCAUUUGCAGCUCUUUCACAGGGCG .(((((((.((......((((((...((((((....)))))).))))))..)))))))))....((......))((....)).(((((.....))))). ( -31.50) >DroSim_CAF1 48078 99 + 1 CGGUGUCAAGAAUCGCAUGUUGAGAAGUCUGCCCAGGCGGAUGUCAAUAAAUCUGACACCCAACGAAAUCCGCCACAUUUGCAGCUCUUUCACAGGGCG .(((((((.((......((((((...((((((....)))))).))))))..)))))))))...........((.......)).(((((.....))))). ( -29.00) >DroEre_CAF1 41759 80 + 1 CGGUGUCAAGAAUCGCAUAUUGAGAAGUCUGCCCAGGCGGAUGUCAAUAAGUCUGGCACUCAAAACAAGCC-------------------CAGAGGGCG .(((((((.((......((((((...((((((....)))))).))))))..)))))))))........(((-------------------(...)))). ( -27.50) >DroYak_CAF1 46946 99 + 1 CGGUGUCAAGAAUCGCAUAUUGAGAAGUCUGCCCAGGCGGAUGUCAAUAAAUCUGACACUCAACACAAUCCGCCACGAUUUGAGCUCUUUCACAGGGCG .(((((((.((......((((((...((((((....)))))).))))))..)))))))))...((.((((......)))))).(((((.....))))). ( -27.00) >consensus CGGUGUCAAGAAUCGCAUAUUGAGAAGUCUGCCCAGGCGGAUGUCAAUAAAUCUGACACCCAACACAAUCCGCCACAUUUGCAGCUCUUUCACAGGGCG .(((((((.((......((((((...((((((....)))))).))))))..))))))))).......................(((((.....))))). (-25.16 = -25.52 + 0.36)

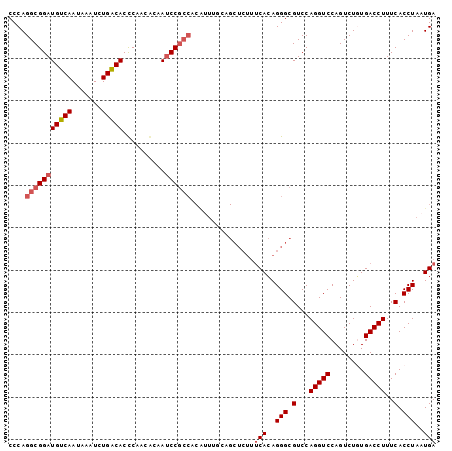
| Location | 6,789,048 – 6,789,149 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -26.42 |
| Consensus MFE | -20.54 |
| Energy contribution | -21.38 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.78 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815912 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6789048 101 + 20766785 CCCAGGCGGAUGUCAAUAAAUCUGACACCCAACACAAUCCGCCACAUUUUCAGCUCUUUCACAGGGCGUCCAGGUCCAGUCUGUGACCUUUCACCUAAUGA ....(((((((((((.......)))))..........)))))).((((....(((((.....)))))(((((((.....)))).))).........)))). ( -26.30) >DroSec_CAF1 39196 101 + 1 CCCAGGCGGAUGUCAAUAAAUCUGACACCCAACGGAAUCCGCCGCAUUUGCAGCUCUUUCACAGGGCGUCCAGGUCCAGUCUGUGACCUUUCACCUAAUGA ....(((((((((((.......)))).((....)).)))))))((....)).(((((.....)))))(((((((.....)))).))).............. ( -29.70) >DroSim_CAF1 48109 101 + 1 CCCAGGCGGAUGUCAAUAAAUCUGACACCCAACGAAAUCCGCCACAUUUGCAGCUCUUUCACAGGGCGUCCAGGUCCAGUCUGUGACCUUUCACCUAAUGA ....(((((((((((.......)))))..........)))))).((((....(((((.....)))))(((((((.....)))).))).........)))). ( -26.30) >DroEre_CAF1 41790 82 + 1 CCCAGGCGGAUGUCAAUAAGUCUGGCACUCAAAACAAGCC-------------------CAGAGGGCGUCCAGGUCCAGUCUUUGACCUUUCACCUAAUGA ...(((.(((.(((((..((.((((.(((........(((-------------------(...)))).....))))))).)))))))..))).)))..... ( -24.42) >DroYak_CAF1 46977 101 + 1 CCCAGGCGGAUGUCAAUAAAUCUGACACUCAACACAAUCCGCCACGAUUUGAGCUCUUUCACAGGGCGUCCAGGUCCAGUCUUUGACCUUUCACCUAAUGA ....(((((((((((.......)))))..........))))))......((((((((.....)))))....(((((........))))).)))........ ( -25.40) >consensus CCCAGGCGGAUGUCAAUAAAUCUGACACCCAACACAAUCCGCCACAUUUGCAGCUCUUUCACAGGGCGUCCAGGUCCAGUCUGUGACCUUUCACCUAAUGA ....(((((((((((.......)))))..........))))))...............(((..(((.(...(((((........)))))..).)))..))) (-20.54 = -21.38 + 0.84)

| Location | 6,789,048 – 6,789,149 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 86.44 |
| Mean single sequence MFE | -33.32 |
| Consensus MFE | -24.53 |
| Energy contribution | -25.45 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.624237 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
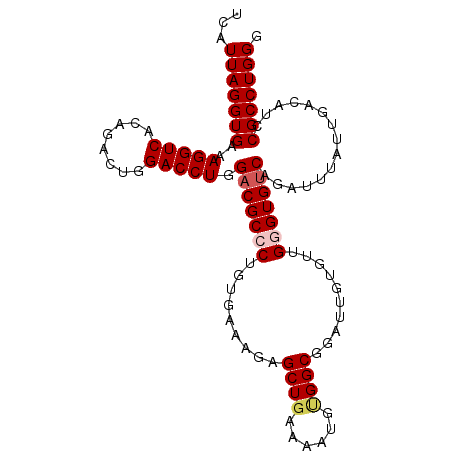
>2R_DroMel_CAF1 6789048 101 - 20766785 UCAUUAGGUGAAAGGUCACAGACUGGACCUGGACGCCCUGUGAAAGAGCUGAAAAUGUGGCGGAUUGUGUUGGGUGUCAGAUUUAUUGACAUCCGCCUGGG ...(((((((..(((((........))))).(((((((.....((..(((........)))...)).....)))))))...............))))))). ( -30.60) >DroSec_CAF1 39196 101 - 1 UCAUUAGGUGAAAGGUCACAGACUGGACCUGGACGCCCUGUGAAAGAGCUGCAAAUGCGGCGGAUUCCGUUGGGUGUCAGAUUUAUUGACAUCCGCCUGGG ((((..((((..(((((........)))))...))))..))))....(((((....))))).....(((.((((((((((.....))))))))))..))). ( -38.20) >DroSim_CAF1 48109 101 - 1 UCAUUAGGUGAAAGGUCACAGACUGGACCUGGACGCCCUGUGAAAGAGCUGCAAAUGUGGCGGAUUUCGUUGGGUGUCAGAUUUAUUGACAUCCGCCUGGG ...(((((((..(((((........))))).(((((((..(((((..((..(....)..))...)))))..)))))))...............))))))). ( -39.50) >DroEre_CAF1 41790 82 - 1 UCAUUAGGUGAAAGGUCAAAGACUGGACCUGGACGCCCUCUG-------------------GGCUUGUUUUGAGUGCCAGACUUAUUGACAUCCGCCUGGG ...(((((((....(((((((.(((((((..(((((((...)-------------------)))..)))..).)).)))).))..)))))...))))))). ( -27.50) >DroYak_CAF1 46977 101 - 1 UCAUUAGGUGAAAGGUCAAAGACUGGACCUGGACGCCCUGUGAAAGAGCUCAAAUCGUGGCGGAUUGUGUUGAGUGUCAGAUUUAUUGACAUCCGCCUGGG ((((..((((..(((((........)))))...))))..))))...........(((.(((((((.((..(((((.....)))))...)))))))))))). ( -30.80) >consensus UCAUUAGGUGAAAGGUCACAGACUGGACCUGGACGCCCUGUGAAAGAGCUGAAAAUGUGGCGGAUUGUGUUGGGUGUCAGAUUUAUUGACAUCCGCCUGGG ...(((((((..(((((........))))).(((((((.........((((......))))..........)))))))...............))))))). (-24.53 = -25.45 + 0.92)

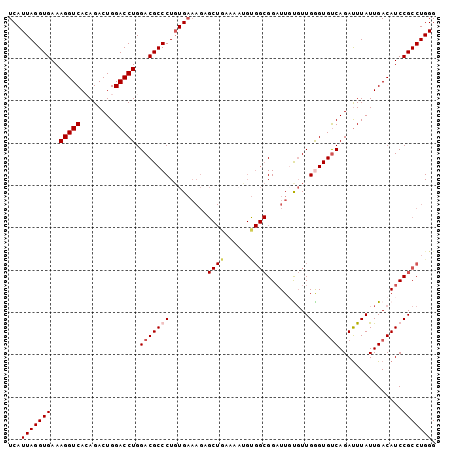
| Location | 6,789,116 – 6,789,219 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 95.73 |
| Mean single sequence MFE | -32.16 |
| Consensus MFE | -26.72 |
| Energy contribution | -27.44 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972047 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6789116 103 - 20766785 UUCGGCCGGAGGGUGUCGAUAUUAGUGGUAUUUAUUACCGCUUCGAAUCCAGUUUUCAGUUUUUACCCAAUCAUUAGGUGAAAGGUCACAGACUGGACCUGGA .....((((......((((....(((((((.....))))))))))).((((((((..(.((((((((.........)))))))).)...)))))))).)))). ( -32.10) >DroSec_CAF1 39264 102 - 1 UUCG-CCGGAGGGUGUCGAUAUUAGUAAUAUUUAUUACCGCUUCAAAUCCAGUUUUCAGUUUUUACCCAAUCAUUAGGUGAAAGGUCACAGACUGGACCUGGA ....-((((((((((..(((((.....)))))...)))).))))...((((((((..(.((((((((.........)))))))).)...))))))))...)). ( -26.90) >DroSim_CAF1 48177 103 - 1 UUCGGCCGGAGGGUGUCGAUAUUAGUGGUAUUUAUUACCGCUUCGAAUCCAGUUUUCAGUUUUUACCCAAUCAUUAGGUGAAAGGUCACAGACUGGACCUGGA .....((((......((((....(((((((.....))))))))))).((((((((..(.((((((((.........)))))))).)...)))))))).)))). ( -32.10) >DroEre_CAF1 41839 103 - 1 UUCGGCCGGAGGGUGUCGAUAUUAGUGGUAUUUAUUACCGCUUCGAAUCCAGUUUUCAGCUUUUAGCCAAUCAUUAGGUGAAAGGUCAAAGACUGGACCUGGA .....((((......((((....(((((((.....))))))))))).(((((((((..((((((.(((........))).))))))..))))))))).)))). ( -36.60) >DroYak_CAF1 47045 103 - 1 UUCGGCCGGAGGGUGUCGAUAUUAGUGGUAUUUAUUGCCGCUUCAGAGCCAGUUUUCAGUUUUUACCCAAUCAUUAGGUGAAAGGUCAAAGACUGGACCUGGA .((((((.....).)))))....(((((((.....)))))))((((..((((((((.(.((((((((.........)))))))).)..))))))))..)))). ( -33.10) >consensus UUCGGCCGGAGGGUGUCGAUAUUAGUGGUAUUUAUUACCGCUUCGAAUCCAGUUUUCAGUUUUUACCCAAUCAUUAGGUGAAAGGUCACAGACUGGACCUGGA .....((((......((((....(((((((.....))))))))))).((((((((..(.((((((((.........)))))))).)...)))))))).)))). (-26.72 = -27.44 + 0.72)


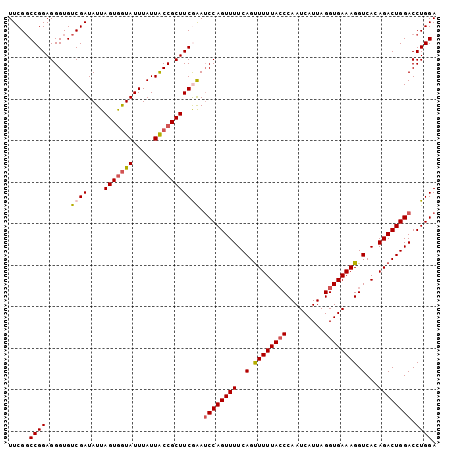
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:42 2006