| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,754,252 – 6,754,384 |
| Length | 132 |
| Max. P | 0.773836 |

| Location | 6,754,252 – 6,754,344 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 92 |
| Reading direction | reverse |
| Mean pairwise identity | 80.80 |
| Mean single sequence MFE | -25.08 |
| Consensus MFE | -15.06 |
| Energy contribution | -16.65 |
| Covariance contribution | 1.59 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.60 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6754252 92 - 20766785 ACUUUCAAUCGCUGAACAAUAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGCGGCAGUUCCGGAAUAUAUCCAGCACCUCAAGGAUUC .....(((((.(((((((.....(((.....)))...))))))).)))))((((((((.....)))........))))).((....)).... ( -27.10) >DroSec_CAF1 5190 92 - 1 ACUUCCAAUCGCUGAACAAUAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGCGGCAGUUCCAGACUAUAUCCAGCACCCCAAGGAUUC .....(((((.(((((((.....(((.....)))...))))))).)))))(((((....((((...))))....))))).((....)).... ( -27.10) >DroSim_CAF1 7893 92 - 1 ACUUCCAAUCGCUGAACAACAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGUGGCAGCUCCGGACUAUAUCCAGCACCUCAAGGAUUU (((.((((((.(((((((.....(((.....)))...))))))).))))))..)))....(((..(((.....)))))).((....)).... ( -27.20) >DroEre_CAF1 5043 92 - 1 ACUUCCAAUCGGUGAACCAUAUUGUCAAUAUGGCCAACGUUCAGGGAUUGGCUGGCGGCAAUUCCGGAAUUUAUCCAGCACCUCAAGGAUUC ...(((....((((..((((((.....)))))).....((((.(((((((.(....).))))))).))))........))))....)))... ( -24.80) >DroYak_CAF1 7760 92 - 1 ACUUCCAAUCGCUGAAUAAUAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGACUGGCGGCAGUACCGCAUCAUAUCCAGCACCUCAAGGAUUC .....(((((.(((((((.....(((.....)))...))))))).)))))....((((.....)))).....((((..........)))).. ( -23.60) >DroPer_CAF1 5606 86 - 1 ACUUCUCGGCUCUGAACAACAUCGUGAACAUGGCCAAUGUCCAAGGACUGGGAGGAUCUAACCAAGGAUUCUAUCAAAAUCCCCAG------ .((((((((((.((.((......))...)).))))...(((....))).))))))..........(((((.......)))))....------ ( -20.70) >consensus ACUUCCAAUCGCUGAACAAUAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGCGGCAGUUCCGGAAUAUAUCCAGCACCUCAAGGAUUC .....(((((.(((((((.....(((.....)))...))))))).)))))((((((((.....)))........)))))............. (-15.06 = -16.65 + 1.59)

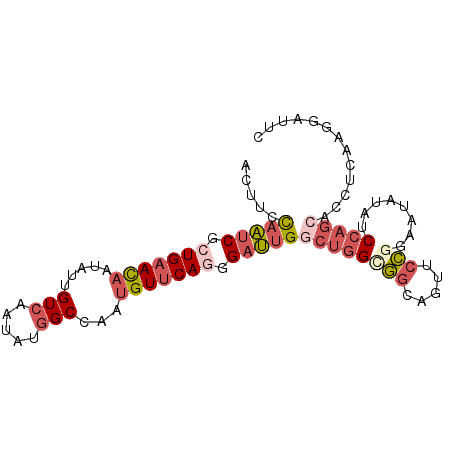

| Location | 6,754,280 – 6,754,384 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 79.94 |
| Mean single sequence MFE | -34.58 |
| Consensus MFE | -20.24 |
| Energy contribution | -21.72 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.773836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6754280 104 - 20766785 CAGCUGCCUAAUGGAUUUGUGCCCAAUCCAGCCGUAGAUAACUUUCAAUCGCUGAACAAUAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGCGGCAGUUC .(((((((...(((........)))..((((((...((......))((((.(((((((.....(((.....)))...))))))).)))))))))).))))))). ( -39.10) >DroSec_CAF1 5218 104 - 1 CAGCUGCCCAAUGGAUUUGUGCCCAAUCCUGCCGUAGAUAACUUCCAAUCGCUGAACAAUAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGCGGCAGUUC .(((((((....(((((.......))))).(((((.....))..((((((.(((((((.....(((.....)))...))))))).))))))..)))))))))). ( -37.60) >DroSim_CAF1 7921 104 - 1 CAGCUGCCCAAUGGAUUUGUGCCCAAUCCUGCCGUGGACAACUUCCAAUCGCUGAACAACAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGUGGCAGCUC .(((((((...(((........))).(((......)))..(((.((((((.(((((((.....(((.....)))...))))))).))))))..)))))))))). ( -38.80) >DroYak_CAF1 7788 104 - 1 CAGCUGCCUAAUGGAUUUGUGCCUAACCCGGCCGUAGACAACUUCCAAUCGCUGAAUAAUAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGACUGGCGGCAGUAC ..((((((...((..(.((.(((......))))).)..)).....(((((.(((((((.....(((.....)))...))))))).)))))...))))))..... ( -33.50) >DroAna_CAF1 5842 98 - 1 CAACUGCCCAACGGAUUUGUACCACAGCCUUCGGUGGAUAACUUCCAGGCUCUGAACAGCAUCGUGAACAUGGCCAAUUCCCAGGGACUGGGCGGCGG------ ...(((((....(((.((((.((((........))))))))..))).((((.((.((......))...)).))))....(((((...))))).)))))------ ( -29.70) >DroPer_CAF1 5628 104 - 1 CAGCUGCCCAACGGAUUCGUGCCCCAGCCAUCGGUGGACAACUUCUCGGCUCUGAACAACAUCGUGAACAUGGCCAAUGUCCAAGGACUGGGAGGAUCUAACCA ..((((....(((....)))....))))....((((((...((((((((((.((.((......))...)).))))...(((....))).)))))).))).))). ( -28.80) >consensus CAGCUGCCCAAUGGAUUUGUGCCCAAUCCUGCCGUAGACAACUUCCAAUCGCUGAACAACAUUGUCAAUAUGGCCAAUGUUCAGGGAUUGGCUGGCGGCAGUUC ..((((((....((.............))...............((((((.(((((((.....(((.....)))...))))))).))))))..))))))..... (-20.24 = -21.72 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:30 2006