
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,695,287 – 6,695,388 |
| Length | 101 |
| Max. P | 0.703648 |

| Location | 6,695,287 – 6,695,388 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -34.73 |
| Consensus MFE | -26.70 |
| Energy contribution | -26.37 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.703648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6695287 101 + 20766785 UGUACGA-GGUUGUUGCUGUGGCUGAAAGCAGUUCACUAGCAACAACAAGGG---------CCACAGCAACAGUGAGUGAGUGACGGCAACGACCGUGAAAGGCCAUUUGC .......-..((((((((((((((....((((....)).))..(.....)))---------))))))))))))...(..((((.((....)).((......)).))))..) ( -33.90) >DroSec_CAF1 43896 101 + 1 UGUACGA-GGUUGUUGCUGUGGCUGAAAGCAGUUCCUUAGCAACAACAAGGA---------CCACAGCAACAGUGAGUGAGUGACGGCAACGACCGUGAAAGGCCAUUUGC .......-..(((((((((((((((....))).(((((.(......))))))---------))))))))))))...(..((((.((....)).((......)).))))..) ( -32.60) >DroSim_CAF1 49235 101 + 1 UGUACGA-GGUUGUUGCUGGGGCUGAGAGCAGUUUCCUAGCAACAACAAGGA---------CCACAGCAACAGUGAGUGAGUGACGGGAACGACCGUGAAAGGCCAUUUGC .((.(..-.((((((((((((((((....))))..))))))))))))..).)---------)....((((..(((.((...(.((((......)))).)...))))))))) ( -34.50) >DroEre_CAF1 45636 102 + 1 UGUACAGAGGUUGUUGCUGCGGCUGAAAGCAGUUCAGCAGCAACAACAAGGG---------CCACAGCAACAGUAAGUGAGUGACGGCAACGACCGUGAAAGGCCAUUUGC ....((((.(((((((((((.((((....))))...)))))))))))...((---------((.((...((.....))...))((((......))))....)))).)))). ( -36.80) >DroYak_CAF1 44234 101 + 1 UGUACCGA-GUUGUUGCUGUGGCUGAAAGCAGUUCAGCAGCAACAACAAGGG---------CCACAGCAACAGUGAGUGAGUGACGGCAACGACCGUGAAAGGCCAUUUGC ((..((..-(((((((((((.((((....))))...)))))))))))..)).---------.))..((((..(((.((...(.((((......)))).)...))))))))) ( -35.80) >DroPer_CAF1 69270 102 + 1 ---------GUUGUUGCCGGGGCAGAAAACUGUUCAACAGCGCCAACAAAGCAGGCAGAGGCAGAGGCAACGACGAGUGAGUGACGGCAACGACCGUGAAAGGCCAUUUGC ---------(((((((((.((((((....))))))......(((.........))).........)))))))))..(..((((.((....)).((......)).))))..) ( -34.80) >consensus UGUACGA_GGUUGUUGCUGUGGCUGAAAGCAGUUCACCAGCAACAACAAGGG_________CCACAGCAACAGUGAGUGAGUGACGGCAACGACCGUGAAAGGCCAUUUGC .........((((((((((.(((((....)))))...)))))))))).............................(..((((.((....)).((......)).))))..) (-26.70 = -26.37 + -0.33)

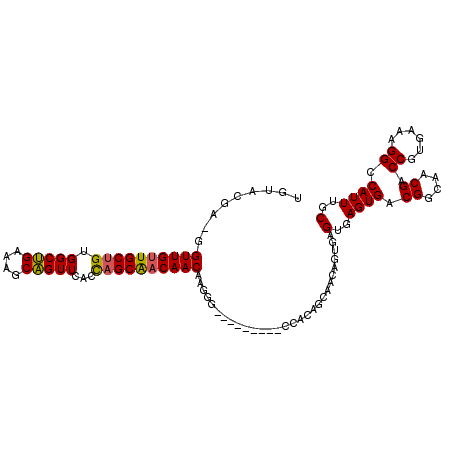
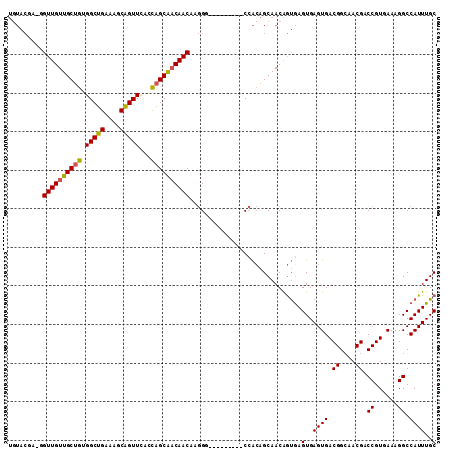
| Location | 6,695,287 – 6,695,388 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 83.99 |
| Mean single sequence MFE | -32.90 |
| Consensus MFE | -23.04 |
| Energy contribution | -23.40 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.54 |
| Structure conservation index | 0.70 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.692762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6695287 101 - 20766785 GCAAAUGGCCUUUCACGGUCGUUGCCGUCACUCACUCACUGUUGCUGUGG---------CCCUUGUUGUUGCUAGUGAACUGCUUUCAGCCACAGCAACAACC-UCGUACA ((((.(((((......)))))))))..............(((((((((((---------(...((.....((.((....))))...))))))))))))))...-....... ( -33.90) >DroSec_CAF1 43896 101 - 1 GCAAAUGGCCUUUCACGGUCGUUGCCGUCACUCACUCACUGUUGCUGUGG---------UCCUUGUUGUUGCUAAGGAACUGCUUUCAGCCACAGCAACAACC-UCGUACA ((((.(((((......)))))))))..............(((((((((((---------(((((((....)).))))).(((....))))))))))))))...-....... ( -33.80) >DroSim_CAF1 49235 101 - 1 GCAAAUGGCCUUUCACGGUCGUUCCCGUCACUCACUCACUGUUGCUGUGG---------UCCUUGUUGUUGCUAGGAAACUGCUCUCAGCCCCAGCAACAACC-UCGUACA (..(((((((......)))))))..).............((((((((.((---------.....((((..((.((....))))...))))))))))))))...-....... ( -29.30) >DroEre_CAF1 45636 102 - 1 GCAAAUGGCCUUUCACGGUCGUUGCCGUCACUCACUUACUGUUGCUGUGG---------CCCUUGUUGUUGCUGCUGAACUGCUUUCAGCCGCAGCAACAACCUCUGUACA ((((.(((((......))))))))).(((((..((.....))....))))---------)..((((((((((.((((((.....)))))).)))))))))).......... ( -36.50) >DroYak_CAF1 44234 101 - 1 GCAAAUGGCCUUUCACGGUCGUUGCCGUCACUCACUCACUGUUGCUGUGG---------CCCUUGUUGUUGCUGCUGAACUGCUUUCAGCCACAGCAACAAC-UCGGUACA ((((.(((((......))))))))).(((((..((.....))....))))---------)((..(((((((((((((((.....))))))...)))))))))-..)).... ( -35.90) >DroPer_CAF1 69270 102 - 1 GCAAAUGGCCUUUCACGGUCGUUGCCGUCACUCACUCGUCGUUGCCUCUGCCUCUGCCUGCUUUGUUGGCGCUGUUGAACAGUUUUCUGCCCCGGCAACAAC--------- ((((.(((((......)))))))))...............((((((...((....(((.((...)).)))((((.....)))).....))...))))))...--------- ( -28.00) >consensus GCAAAUGGCCUUUCACGGUCGUUGCCGUCACUCACUCACUGUUGCUGUGG_________CCCUUGUUGUUGCUACUGAACUGCUUUCAGCCACAGCAACAACC_UCGUACA ((((.(((((......)))))))))..............(((((((((((..............((....)).......(((....))))))))))))))........... (-23.04 = -23.40 + 0.36)
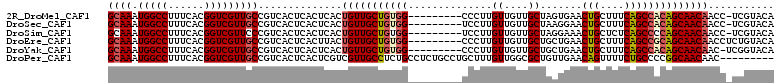
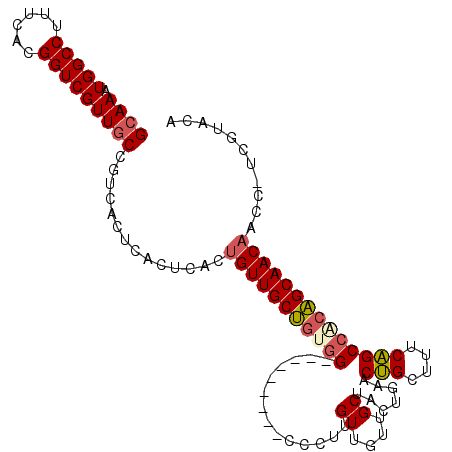
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:09 2006