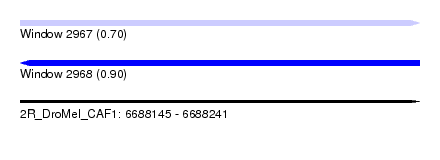
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,688,145 – 6,688,241 |
| Length | 96 |
| Max. P | 0.901214 |

| Location | 6,688,145 – 6,688,241 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -37.23 |
| Consensus MFE | -28.15 |
| Energy contribution | -28.48 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.704581 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
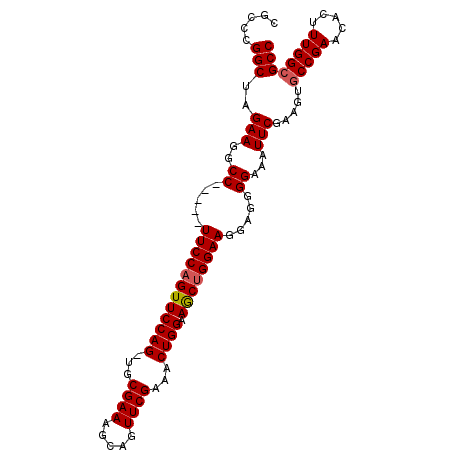
>2R_DroMel_CAF1 6688145 96 + 20766785 GGCGCCAAAGUGUUCGGCACCACGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCA-CUGGAACUGGAA-----GGCCUUCUAGCCGGGCG ..((((...(((((((......))))).......((((((((.(((((((..((((.....)))).)-))))))))))))-----)).......))..)))) ( -35.80) >DroSec_CAF1 36686 96 + 1 GGCGCCAAAGUGUUCGGCACUUCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCA-CUGGAACUGGAA-----GGCCUUCUAGCCGGGCG ...........(((((((.....(.....)....((((((((.(((((((..((((.....)))).)-))))))))))))-----)).......))))))). ( -35.60) >DroSim_CAF1 40340 96 + 1 GGCGCCAAAGUGUUCGGCACUUCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCA-CUGGAACUGGAA-----GGCCUUCUAGCCGGGCG ...........(((((((.....(.....)....((((((((.(((((((..((((.....)))).)-))))))))))))-----)).......))))))). ( -35.60) >DroEre_CAF1 34234 101 + 1 GGCGCCAAAGUGUUCGGCACCUCGAAUUUCCCCUACUUCCCGCUUCCAGUUUCGAACUGCUUUCGCA-CUGGAACUGGAAGCAGUGGCUUUCUAGCCGGCUG ...(((.....((((((....))))))..............(((((((((((((...(((....)))-.)))))))))))))...((((....))))))).. ( -36.70) >DroYak_CAF1 36788 100 + 1 GGCGCCAAAGUUUUCGGAAACCCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCA-CUGGAACUGGAAGGAA-GGCCUUCUAGCCGGCUA ...(((......(((((....))))).......(((((((((.(((((((..((((.....)))).)-))))))))))))))).-(((......)))))).. ( -42.30) >DroAna_CAF1 35500 92 + 1 GGCGCCAAACUGUUCGGCUGUUCGAAUUUCCCCUCCUUCCAGUUUCCAGU-GCGAA----UUUCGCGCCUGGAACUGGAA-----GGCCUUCUAGCCGGCAA ...(((.....((((((....)))))).......((((((((((.(((((-(((..----...)))).))))))))))))-----))..........))).. ( -37.40) >consensus GGCGCCAAAGUGUUCGGCACCUCGAAUUUCCCCUCCUUCCAGCUUCCAGUUUCGAACUGCUUUCGCA_CUGGAACUGGAA_____GGCCUUCUAGCCGGCCG ((((((.........))).....(((...((.....((((((.((((((...((((.....))))...)))))))))))).....))..)))..)))..... (-28.15 = -28.48 + 0.33)

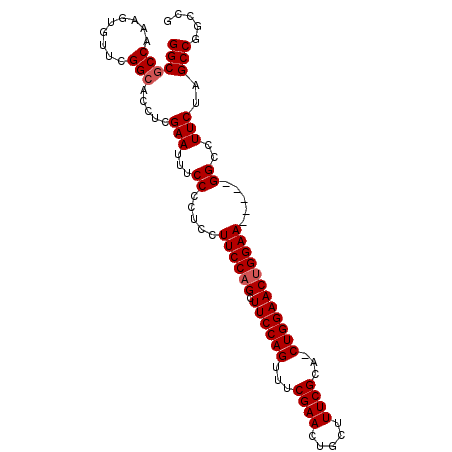
| Location | 6,688,145 – 6,688,241 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 88.29 |
| Mean single sequence MFE | -42.25 |
| Consensus MFE | -32.32 |
| Energy contribution | -32.52 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -3.62 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.02 |
| SVM RNA-class probability | 0.901214 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
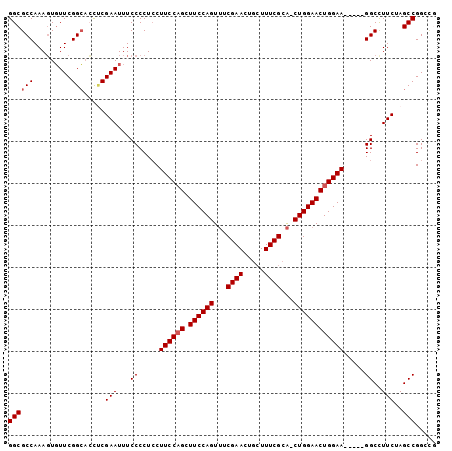
>2R_DroMel_CAF1 6688145 96 - 20766785 CGCCCGGCUAGAAGGCC-----UUCCAGUUCCAG-UGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGUGGUGCCGAACACUUUGGCGCC ..(((((((....))))-----((((((((((((-(.((((.....))))..))))).))))))))...))).........((((((((.....)))))))) ( -42.40) >DroSec_CAF1 36686 96 - 1 CGCCCGGCUAGAAGGCC-----UUCCAGUUCCAG-UGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGAAGUGCCGAACACUUUGGCGCC ..(((((((....))))-----((((((((((((-(.((((.....))))..))))).))))))))...)))..........(((((((.....))))))). ( -40.10) >DroSim_CAF1 40340 96 - 1 CGCCCGGCUAGAAGGCC-----UUCCAGUUCCAG-UGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGAAGUGCCGAACACUUUGGCGCC ..(((((((....))))-----((((((((((((-(.((((.....))))..))))).))))))))...)))..........(((((((.....))))))). ( -40.10) >DroEre_CAF1 34234 101 - 1 CAGCCGGCUAGAAAGCCACUGCUUCCAGUUCCAG-UGCGAAAGCAGUUCGAAACUGGAAGCGGGAAGUAGGGGAAAUUCGAGGUGCCGAACACUUUGGCGCC ...((((((....)))).(((((((((((((.(.-(((....))).)..).))))))))))))......))..........((((((((.....)))))))) ( -45.40) >DroYak_CAF1 36788 100 - 1 UAGCCGGCUAGAAGGCC-UUCCUUCCAGUUCCAG-UGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGGGUUUCCGAAAACUUUGGCGCC .....(((..((((.((-((((((((((((((((-(.((((.....))))..))))).))))))))))))))....(((((....)))))..))))...))) ( -47.20) >DroAna_CAF1 35500 92 - 1 UUGCCGGCUAGAAGGCC-----UUCCAGUUCCAGGCGCGAAA----UUCGC-ACUGGAAACUGGAAGGAGGGGAAAUUCGAACAGCCGAACAGUUUGGCGCC .....(((.......((-----((((((((((((..(((...----..)))-.)))).))))))))))................(((((.....)))))))) ( -38.30) >consensus CGCCCGGCUAGAAGGCC_____UUCCAGUUCCAG_UGCGAAAGCAGUUCGAAACUGGAAGCUGGAAGGAGGGGAAAUUCGAAGUGCCGAACACUUUGGCGCC .....(((..(((..((.....((((((((((((...((((.....))))...)))).)))))))).....))...))).....(((((.....)))))))) (-32.32 = -32.52 + 0.19)

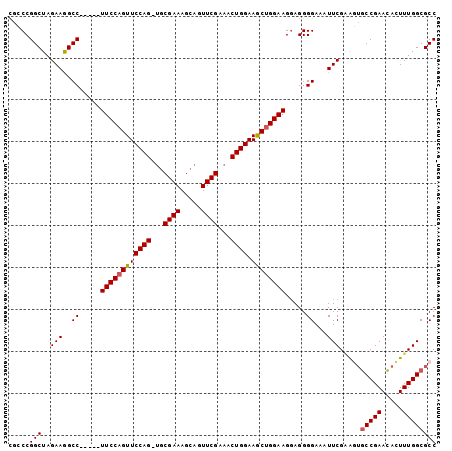
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:06 2006