
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,672,146 – 6,672,250 |
| Length | 104 |
| Max. P | 0.855618 |

| Location | 6,672,146 – 6,672,250 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -33.77 |
| Consensus MFE | -19.45 |
| Energy contribution | -21.37 |
| Covariance contribution | 1.92 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.81 |
| SVM RNA-class probability | 0.855618 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6672146 104 + 20766785 AAGUUUUGCCGCAUUUUU--GGUGGCAGCUGCAGAGCUUAACUAAAGUUGUGUAACUCAACUAAAGUAAAGUCUCUUUAGCCCGGCGAC--------------UUAUCGUGUUGCAGAAG ...((((((.((((....--(((.((.((((.(((((((.(((..(((((.......)))))..))).))).)))).))))...)).))--------------)....)))).)))))). ( -34.50) >DroSec_CAF1 20427 104 + 1 AAGUUUUGCCGCAUUUUU--GGUGGCAGCUGAAGAGCUUAACUAAAGUUGUGUACCUCAACUAAAGUAAAGUCUCUUUAGCCCGGCGAC--------------UUAUCGUGUUGCUGAAG ......((((((......--.))))))((((((((((((.(((..(((((.......)))))..))).))).))))))))).(((((((--------------.......)))))))... ( -39.60) >DroSim_CAF1 22833 104 + 1 AAGUUUUGCCGCAUUUUU--GGUGGCAGCUGCAGAGCUUAACUAAAGUUGUGUAACUCAACUAAAGUAAAGUCUCUUUAGCCCGGCGAC--------------UUAUCGUGUUGCAGAAG ...((((((.((((....--(((.((.((((.(((((((.(((..(((((.......)))))..))).))).)))).))))...)).))--------------)....)))).)))))). ( -34.50) >DroEre_CAF1 19122 104 + 1 AAGUUUUGCCGCAUUUUU--GGCGGCAGAUGCAGAGCUUAACUAAAGUUGUGUAACUAAACUAAAGCAAAGUCACUUUAGCCCAGCGAC--------------UUAUCGUGUUGCAGAAG .(((((((((((......--.))))))))((((.(((((.....))))).)))))))...((((((........))))))....(((((--------------.......)))))..... ( -30.00) >DroYak_CAF1 20706 103 + 1 AAGU-UUGCCGCAUUUUU--GGUGGCAGAUGCAGAGCUUAACUAAAGUUGUGUAACUCAACUAAAGUAAAGUCUCUUUAGCCCUGCGAC--------------UUAUCGUGUUGCAGAAG ..((-(((((((......--.)))))))))(((((((((.(((..(((((.......)))))..))).))).))))...)).(((((((--------------.......)))))))... ( -34.40) >DroAna_CAF1 22106 116 + 1 AAACUUUGCGGCUUCUGGGGGGCGACAGAUGUGGAGAUCAACUAAAGUUGUAGAACUCAUCUGG----AAGUCCCUUCCUCUCCCCAAACUACACACAAAUAUUCUUCGUUUUGCGGAAG ....((((.((.....(((((((..((((((.(.....((((....)))).....).)))))).----..))))))).....)).))))................((((.....)))).. ( -29.60) >consensus AAGUUUUGCCGCAUUUUU__GGUGGCAGAUGCAGAGCUUAACUAAAGUUGUGUAACUCAACUAAAGUAAAGUCUCUUUAGCCCGGCGAC______________UUAUCGUGUUGCAGAAG ......((((((.........))))))((((.(((((((.(((..(((((.......)))))..))).))).)))).)))).(((((((.....................)))))))... (-19.45 = -21.37 + 1.92)

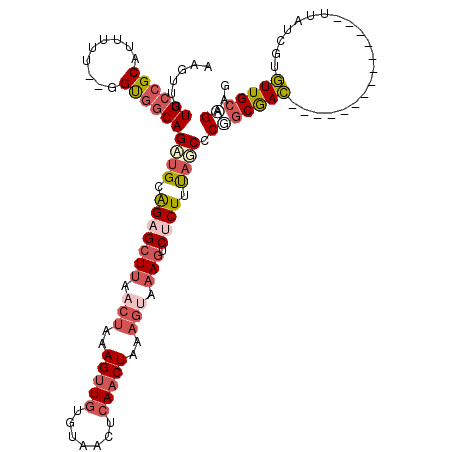
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:01 2006