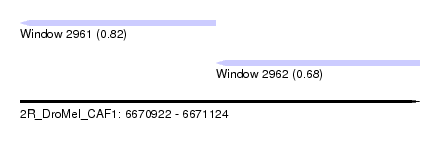
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,670,922 – 6,671,124 |
| Length | 202 |
| Max. P | 0.821718 |

| Location | 6,670,922 – 6,671,021 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 77.87 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -21.87 |
| Energy contribution | -22.23 |
| Covariance contribution | 0.37 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.28 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.68 |
| SVM RNA-class probability | 0.821718 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6670922 99 - 20766785 AAGAAGAAAAACACGGGGUCAAUCCGGUAACAGUUCACGGAAACUAUUGUCCCAACUCACCGAUUAUCUUCUGGAUGGCCGUCCGGGCAUCCAGGAUCU ....(((........(((.(((((((...........)))).....))).)))...............(((((((((.((....)).)))))))))))) ( -29.10) >DroSec_CAF1 19348 99 - 1 AAGAAACACGGAAGGGGGUCAAUCCGGUAACAGUUCACGGAAACUAUUGGCCCAACUCACCGAUUAUCUUCUGGAUGGCCGUCCGGGCGUCCAGGAUCU .(((....(((..(((((((((((((...........)))).....)))))))..))..)))......(((((((((.((....)).)))))))))))) ( -35.20) >DroSim_CAF1 19576 95 - 1 ----AACACGGAAAGGGGUCAAUCCGGUAACAGUUCACGGAAACUAUUGGCACAACUCACCGAUUAUCUUCUGGAUGGCCGUCCGGGCGUCCAGGAUCU ----....(((..((..(((((((((...........)))).....)))))....))..)))......(((((((((.((....)).)))))))))... ( -30.30) >DroEre_CAF1 18235 90 - 1 ----A-----GUUGGGGGUCAAUCCGGUAACAGUUCACGGCAACUAUGGGCCCAACUUACCGAUUAUCUUCUGGAUGGCCGUCCGGGCGUCCAGGAUCU ----.-----.....(((((....(((((..((((...(((........))).)))))))))........(((((((.((....)).)))))))))))) ( -31.40) >DroYak_CAF1 19769 90 - 1 ----A-----ACACGGGGUUAGUCCGGUUACAGUUCACGGAAACUAUUGGGCCAACUCACCGAUUAUCUUCUCGAUGGCCGUCCGGGCGUCCAGAAUCU ----.-----...(((((((.(((((((...((((......))))))))))).))).).)))......((((.((((.((....)).)))).))))... ( -26.20) >DroAna_CAF1 21529 91 - 1 AAGAAUGU-------CAUUAAGUGCUGUAGAUGUCCUCUUAAACUAUUUA-AAAUCUUACCGAUUAUCUUCUGGAUGGCUGUCUGGGCAUCCAGCAGCU ........-------........(((((.(((((((..............-.((((.....)))).......((((....)))))))))))..))))). ( -20.00) >consensus ____A__A__GAACGGGGUCAAUCCGGUAACAGUUCACGGAAACUAUUGGCCCAACUCACCGAUUAUCUUCUGGAUGGCCGUCCGGGCGUCCAGGAUCU ...............(((((((((((...........))).....))))))))...............(((((((((.((....)).)))))))))... (-21.87 = -22.23 + 0.37)


| Location | 6,671,021 – 6,671,124 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -33.80 |
| Consensus MFE | -16.60 |
| Energy contribution | -17.60 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.676521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6671021 103 - 20766785 GGCCACAAAGGUGUUCGGGUCAAUCAAAGUGGCCACCUCCCAGGGGUGAACAACUGAAGGACCCGGAAUCCGAUUGUAAGCGUCUGAAAAA-AGCAAAAAAAAA .((.((((.((..((((((((..(((...((..(((((.....)))))..))..)))..))))))))..))..))))..))..........-............ ( -33.00) >DroSim_CAF1 19671 102 - 1 GGCCACAAAGGUGUUGGGGUCAAUCAAAGUGGCCACCUCCCAGGGGUGAACAACCGAGGGACCCGGAAUCCGAUUGUAUGCGUCUGAAAGA-ACCCAAAAAAU- .((.((((.((..((.(((((..((....((..(((((.....)))))..))...))..))))).))..))..))))..))..........-...........- ( -29.60) >DroEre_CAF1 18325 97 - 1 GGCCACAAAGGUGUUCGGGUCAAUCAAAGUGGCCACCUCCCAGGGGUGAACAACUGAGGGACCCGGAAUGCGGUUGUAUGCGGCUAAAAGA-AAA--GGG---- ((((((((..(((((((((((..(((...((..(((((.....)))))..))..)))..))))).))))))..))))....))))......-...--...---- ( -37.10) >DroYak_CAF1 19859 99 - 1 GGCCACAAAAGUGUUGGGGUCAAUCAAAGUGGCCACCUCCCAGGGGUGAACCACUGAGGGACCCGGAAUCCGGUUGUAUGCGGCUGAAAAA-ACAUAGAA---- ((((((((..(..((.(((((..(((..((((.(((((.....)))))..)))))))..))))).))...)..))))....))))......-........---- ( -34.40) >DroAna_CAF1 21620 100 - 1 GGCCACAAAGGUGUUGGGAUCAAUCACUCGGGCCACUUCCCAGGGAUGAACGACCGAAGGUCCGGGGAUGCGGUUGUAGGCAUCUGGAAGGGAAAUCGAA---- ((((.....(((((((....))).))))..)))).(((((...(((((.(((((((...(((....))).)))))))...))))))))))..........---- ( -35.60) >DroPer_CAF1 46366 93 - 1 GGCCACAAAUGUGUUUGGAUCAAUCACGGUGGCCACCUCCGCAGGGUGGACGACAGAGCGUCCUGGAAUGCGAUUGUAGGCAGCUGCAAAG-AC---------- ((((((....(((.(((...))).))).)))))).....((((....(((((......))))).....)))).((((((....))))))..-..---------- ( -33.10) >consensus GGCCACAAAGGUGUUGGGGUCAAUCAAAGUGGCCACCUCCCAGGGGUGAACAACUGAGGGACCCGGAAUCCGAUUGUAUGCGGCUGAAAAA_ACA_AGAA____ .((.((((..(..((.(((((..((....((..(((((.....)))))..))...))..))))).).)..)..))))..))....................... (-16.60 = -17.60 + 1.00)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:13:00 2006