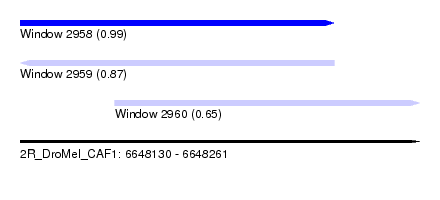
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,648,130 – 6,648,261 |
| Length | 131 |
| Max. P | 0.986551 |

| Location | 6,648,130 – 6,648,233 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -25.93 |
| Consensus MFE | -25.84 |
| Energy contribution | -25.65 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.27 |
| Structure conservation index | 1.00 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
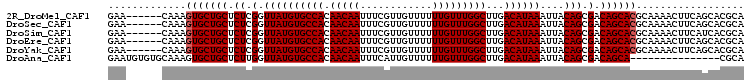
>2R_DroMel_CAF1 6648130 103 + 20766785 ---------UGUUUGUGUUUGUUUUUGCAUAUCUGCAUCCGAACAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACA ---------.(((((((((......((((....)))).((((((((((..........(((.....)))(((((......))))).))))))))))...))))))))).... ( -25.00) >DroSec_CAF1 197506 103 + 1 ---------UGUUUGUGUUUGUUUUUGCAUAUCUGCAUCCGAACAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACA ---------.(((((((((......((((....)))).((((((((((..........(((.....)))(((((......))))).))))))))))...))))))))).... ( -25.00) >DroSim_CAF1 185255 103 + 1 ---------UGUUUGUGUCUGUUUUUGCAUAUCUGCAUCCGAACAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACA ---------.(((((((((......((((....)))).((((((((((..........(((.....)))(((((......))))).))))))))))...))))))))).... ( -27.50) >DroYak_CAF1 205514 112 + 1 UUGUGCUUGCGUUUGUGUUUGUUUUUGCAUAUCUGCAUCCGAACAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACA (((((..((((..(((((........)))))..)))).((((((((((..........(((.....)))(((((......))))).)))))))))).....)))))...... ( -26.20) >consensus _________UGUUUGUGUUUGUUUUUGCAUAUCUGCAUCCGAACAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACA ..........(((((((((......((((....)))).((((((((((..........(((.....)))(((((......))))).))))))))))...))))))))).... (-25.84 = -25.65 + -0.19)

| Location | 6,648,130 – 6,648,233 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 94.88 |
| Mean single sequence MFE | -19.75 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.97 |
| SVM decision value | 0.85 |
| SVM RNA-class probability | 0.867069 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6648130 103 - 20766785 UGUAAUUUAUGUCAAGCCAAACAAAAAACAACGAAAUUGUUGUGGCACAUAACCGAGAGCAGCACUUUGUUCGGAUGCAGAUAUGCAAAAACAAACACAAACA--------- (((...((((((...(((((((((............))))).))))))))))((((..((((....)))))))).((((....))))...)))..........--------- ( -19.20) >DroSec_CAF1 197506 103 - 1 UGUAAUUUAUGUCAAGCCAAACAAAAAACAACGAAAUUGUUGUGGCACAUAACCGAGAGCAGCACUUUGUUCGGAUGCAGAUAUGCAAAAACAAACACAAACA--------- (((...((((((...(((((((((............))))).))))))))))((((..((((....)))))))).((((....))))...)))..........--------- ( -19.20) >DroSim_CAF1 185255 103 - 1 UGUAAUUUAUGUCAAGCCAAACAAAAAACAACGAAAUUGUUGUGGCACAUAACCGAGAGCAGCACUUUGUUCGGAUGCAGAUAUGCAAAAACAGACACAAACA--------- .....(((.((((..(((((((((............))))).))))......((((..((((....)))))))).((((....))))......)))).)))..--------- ( -20.90) >DroYak_CAF1 205514 112 - 1 UGUAAUUUAUGUCAAGCCAAACAAAAAACAACGAAAUUGUUGUGGCACAUAACCGAGAGCAGCACUUUGUUCGGAUGCAGAUAUGCAAAAACAAACACAAACGCAAGCACAA (((...((((((...(((((((((............))))).))))))))))((((..((((....)))))))).((((....))))...)))........(....)..... ( -19.70) >consensus UGUAAUUUAUGUCAAGCCAAACAAAAAACAACGAAAUUGUUGUGGCACAUAACCGAGAGCAGCACUUUGUUCGGAUGCAGAUAUGCAAAAACAAACACAAACA_________ (((...((((((...(((((((((............))))).))))))))))((((..((((....)))))))).((((....))))...)))................... (-19.20 = -19.20 + -0.00)

| Location | 6,648,161 – 6,648,261 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 92.22 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -21.37 |
| Energy contribution | -21.53 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649120 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6648161 100 + 20766785 GAA------CAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACAGCGACAGCACGCAAAACUUCAGCACGCA ...------....((((((.......((((((((((.(((((............)))))))))...))))))......(((......))).......))))))... ( -23.90) >DroSec_CAF1 197537 100 + 1 GAA------CAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACAGCGACAGCACGCAAAACUUCAGCACGCA ...------....((((((.......((((((((((.(((((............)))))))))...))))))......(((......))).......))))))... ( -23.90) >DroSim_CAF1 185286 100 + 1 GAA------CAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACAGCGACAGCACGCAAAACUUCAUCACGCA ...------....((((((.....(((.....))).........((((((((((.((((......)))).)))..)))))))))))))((.............)). ( -23.12) >DroEre_CAF1 198925 100 + 1 GAA------CAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACAGCGACAGCACGCAAAACUUCAGCACGCA ...------....((((((.......((((((((((.(((((............)))))))))...))))))......(((......))).......))))))... ( -23.90) >DroYak_CAF1 205554 100 + 1 GAA------CAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACAGCGACAGCACGCAAAACUUCAGCACGCA ...------....((((((.......((((((((((.(((((............)))))))))...))))))......(((......))).......))))))... ( -23.90) >DroAna_CAF1 212874 91 + 1 GAAUGUGUGCAAAGUGCUGCUCUUGGUUAUGUGCCACAACAAUUUCAUUGUUUUUUGUUUGGCUUGACAUAAAUUACAGCGACAGCA---------------CGCA ...(((((((....(((((.......((((((((((.(((((............)))))))))...))))))....)))))...)))---------------)))) ( -26.80) >consensus GAA______CAAAGUGCUGCUCUCGGUUAUGUGCCACAACAAUUUCGUUGUUUUUUGUUUGGCUUGACAUAAAUUACAGCGACAGCACGCAAAACUUCAGCACGCA .............(((((((.((.(.((((((((((.(((((............)))))))))...))))))....))).).)))))).................. (-21.37 = -21.53 + 0.17)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:58 2006