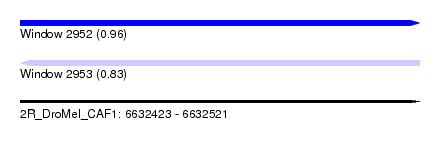
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,632,423 – 6,632,521 |
| Length | 98 |
| Max. P | 0.961721 |

| Location | 6,632,423 – 6,632,521 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -23.50 |
| Consensus MFE | -11.77 |
| Energy contribution | -12.02 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.50 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961721 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
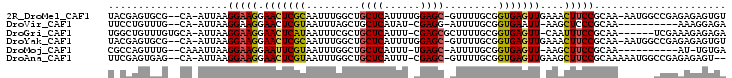
>2R_DroMel_CAF1 6632423 98 + 20766785 ACACUCUCUCGGCCAUU-UUGCGGAAGUUUCAACUCACCGCAAAAC-GCUCCAAAAUGAGCAGCCAAAUUGCGAGUUCCUUCCUUAAU-UG--CGCACUCGUA ..........(((..((-((((((.(((....)))..)))))))).-((((......)))).)))....(((((((.((.........-.)--.).))))))) ( -26.90) >DroVir_CAF1 130290 87 + 1 UCUCCUUU----------UUGCGGGAGCUU-AAUUCACCGCAAAAU-CCUCG-AUAUGAGCAGCUAAAUUACGAGUUCCUUCCUUAAU-UG--CAAACAGGAA ..((((.(----------((((((((((((-........((.....-.(((.-....)))..))........))))))))........-.)--)))).)))). ( -18.19) >DroGri_CAF1 131986 94 + 1 UCUCUCUUUCGA------UUGCGGAAAUUG-AACUCACCGCAAAAGCGCUCG-AAAUGAGCAGCGAAAUUAUGAGUUCCUUCCUUAAU-UGCACAAACAGCCA .........(((------(((.((((...(-((((((..((....))((((.-....))))..........))))))).)))).))))-))............ ( -22.80) >DroYak_CAF1 192429 98 + 1 ACACUCUCUCGGCCAUU-UUGCGGAAGUUUCAACUCACCGCAAAAC-GCUCCAAAAUGAGCAGCCAAAUUGCGAGUUCCUUCCUUAAU-UG--CGCACUCGUA ..........(((..((-((((((.(((....)))..)))))))).-((((......)))).)))....(((((((.((.........-.)--.).))))))) ( -26.90) >DroMoj_CAF1 137209 87 + 1 UCACA-AU----------UUGCGGAAGCUU-AACUCACCGCAAAAU-GCUCA-AAAUGAGCAGCCAAAUUACGAAUUCCUUCCUUAAUUUG--CAAACUGGCG .....-.(----------((((((.((...-..))..))))))).(-((((.-....)))))((((..((.((((((........))))))--.))..)))). ( -23.00) >DroAna_CAF1 199004 96 + 1 --ACUCUCUCGGCCAUUUUUGCGGAAGCUUCAACUCACCGCAAAAC-GCUCG-AAAUGAGCAGCCAAAUUACGAGUUCCUUCCUUAAU-UG--CUCACUCGAA --........(((...((((((((.((......))..)))))))).-((((.-....)))).)))......(((((............-..--...))))).. ( -23.23) >consensus UCACUCUCUCGG______UUGCGGAAGCUU_AACUCACCGCAAAAC_GCUCG_AAAUGAGCAGCCAAAUUACGAGUUCCUUCCUUAAU_UG__CAAACUCGAA ......................(((((....(((((...........((((......))))...........))))).))))).................... (-11.77 = -12.02 + 0.25)



| Location | 6,632,423 – 6,632,521 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 77.85 |
| Mean single sequence MFE | -28.54 |
| Consensus MFE | -15.93 |
| Energy contribution | -15.82 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.833439 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
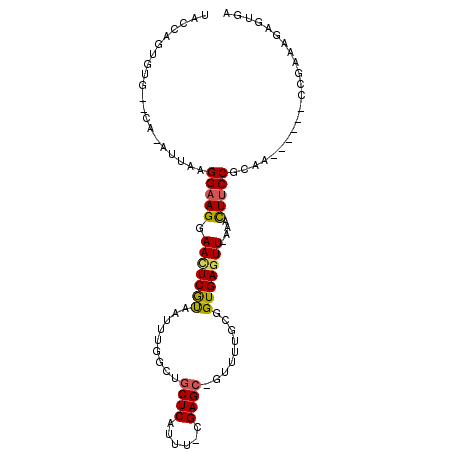
>2R_DroMel_CAF1 6632423 98 - 20766785 UACGAGUGCG--CA-AUUAAGGAAGGAACUCGCAAUUUGGCUGCUCAUUUUGGAGC-GUUUUGCGGUGAGUUGAAACUUCCGCAA-AAUGGCCGAGAGAGUGU ..(((((...--..-............)))))...(((((((((((......))))-(((((((((.((((....))))))))))-))))))))))....... ( -32.23) >DroVir_CAF1 130290 87 - 1 UUCCUGUUUG--CA-AUUAAGGAAGGAACUCGUAAUUUAGCUGCUCAUAU-CGAGG-AUUUUGCGGUGAAUU-AAGCUCCCGCAA----------AAAGGAGA (((((....(--((-.(((((..(.(....).)..))))).)))......-.....-.((((((((.((...-....))))))))----------))))))). ( -20.00) >DroGri_CAF1 131986 94 - 1 UGGCUGUUUGUGCA-AUUAAGGAAGGAACUCAUAAUUUCGCUGCUCAUUU-CGAGCGCUUUUGCGGUGAGUU-CAAUUUCCGCAA------UCGAAAGAGAGA ..(((((....)))-.....((((.((((((((.........((((....-.))))((....)).)))))))-)...))))))..------((....)).... ( -27.80) >DroYak_CAF1 192429 98 - 1 UACGAGUGCG--CA-AUUAAGGAAGGAACUCGCAAUUUGGCUGCUCAUUUUGGAGC-GUUUUGCGGUGAGUUGAAACUUCCGCAA-AAUGGCCGAGAGAGUGU ..(((((...--..-............)))))...(((((((((((......))))-(((((((((.((((....))))))))))-))))))))))....... ( -32.23) >DroMoj_CAF1 137209 87 - 1 CGCCAGUUUG--CAAAUUAAGGAAGGAAUUCGUAAUUUGGCUGCUCAUUU-UGAGC-AUUUUGCGGUGAGUU-AAGCUUCCGCAA----------AU-UGUGA .(((((.(((--(.((((........)))).)))).)))))(((((....-.))))-).(((((((.(((..-...)))))))))----------).-..... ( -27.80) >DroAna_CAF1 199004 96 - 1 UUCGAGUGAG--CA-AUUAAGGAAGGAACUCGUAAUUUGGCUGCUCAUUU-CGAGC-GUUUUGCGGUGAGUUGAAGCUUCCGCAAAAAUGGCCGAGAGAGU-- (((...(((.--..-.))).)))....((((....(((((((((((....-.))))-.((((((((.((((....))))))))))))..))))))).))))-- ( -31.20) >consensus UACCAGUGUG__CA_AUUAAGGAAGGAACUCGUAAUUUGGCUGCUCAUUU_CGAGC_GUUUUGCGGUGAGUU_AAACUUCCGCAA______CCGAAAGAGUGA ....................(((((.(((((((.........((((......)))).........)))))))....)))))...................... (-15.93 = -15.82 + -0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:51 2006