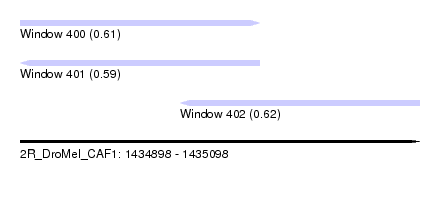
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,434,898 – 1,435,098 |
| Length | 200 |
| Max. P | 0.619050 |

| Location | 1,434,898 – 1,435,018 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -30.90 |
| Consensus MFE | -26.94 |
| Energy contribution | -26.88 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.614153 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1434898 120 + 20766785 CACUGUGGCCUCAACGUUAGUUGACGGCAGCAAUACUCUUGUCUCAGACGUUCCUUUAAACUCGAACGAAACAAAGGAUGAGUUGGAAUUCCUUAUAUCCGAUGUUAUCCAGCAAAUAAC ...(((.((((((((....))))).))).)))......((((......(((((..........))))).......((((((((((((..........)))))).)))))).))))..... ( -29.60) >DroSec_CAF1 52077 120 + 1 CACUGUGGCCUCAACGUUAGUUGACGGCAGCAAUACUCUUGUCUCAGACGUUCCUUUAAACUCGAACGAAACAAAGGAUGAGCUGGAAUUCCUUAUAUCCGAUGUUAUCCAGCAAAUAAC ...(((.((((((((....))))).))).)))......((((......(((((..........))))).((((..(((((....((....))...)))))..)))).....))))..... ( -27.80) >DroEre_CAF1 51438 120 + 1 CACUGUGGCCUCAACGUUAGUUGACGGCAGCAAUUCUCUUGUCUCAGAUGUUCCUUUAAAUUCGAACGAGACGAAGGAUGAGCUGGAAUUCCUCAUAUCCGAUGUUAUCCAGCAAAUAAC ...(((.((((((((....))))).))).)))(((((.(((((((....((((..........))))))))))))))))..((((((...(.((......)).)...))))))....... ( -35.40) >DroYak_CAF1 56474 120 + 1 CACUGUGGCCUCAACGUUAGUUGACGGCAGUAAUAAUCUUGUCUCAGAUGUUCCUUUAAAUUCGAACGAGACAAAGGAUGAGCUGGAAUUCCUUAUAUCCGAUGUUAUUCAGCAAAUAAC ..(((..((((((((....))))).)))(((((((.(((((((((....((((..........))))))))))).(((((....((....))...)))))))))))))))))........ ( -30.80) >consensus CACUGUGGCCUCAACGUUAGUUGACGGCAGCAAUACUCUUGUCUCAGACGUUCCUUUAAACUCGAACGAAACAAAGGAUGAGCUGGAAUUCCUUAUAUCCGAUGUUAUCCAGCAAAUAAC .......((((((((....))))).))).((.......(((((((....((((..........))))))))))).((((((..((((..........))))...)))))).))....... (-26.94 = -26.88 + -0.06)

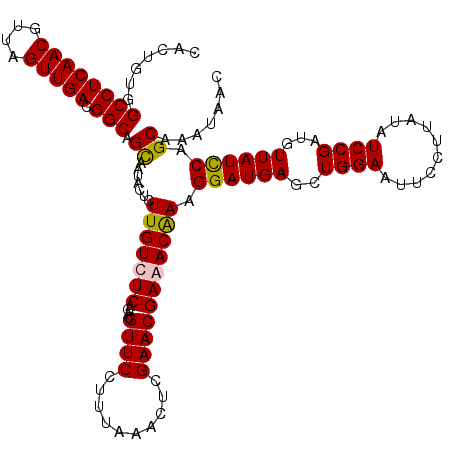
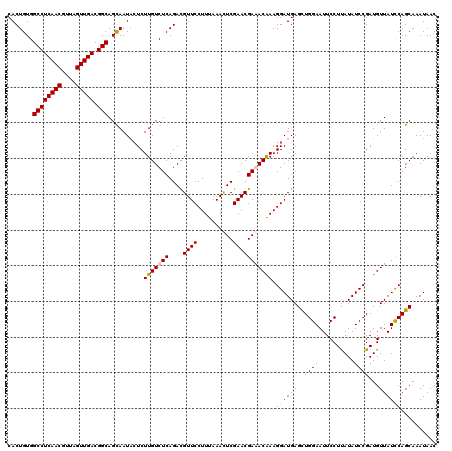
| Location | 1,434,898 – 1,435,018 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.42 |
| Mean single sequence MFE | -33.05 |
| Consensus MFE | -30.24 |
| Energy contribution | -30.30 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.50 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.588139 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1434898 120 - 20766785 GUUAUUUGCUGGAUAACAUCGGAUAUAAGGAAUUCCAACUCAUCCUUUGUUUCGUUCGAGUUUAAAGGAACGUCUGAGACAAGAGUAUUGCUGCCGUCAACUAACGUUGAGGCCACAGUG ((((((.....)))))).((((((....((....))......(((((((.(((....)))..)))))))..))))))........(((((..(((.(((((....))))))))..))))) ( -33.30) >DroSec_CAF1 52077 120 - 1 GUUAUUUGCUGGAUAACAUCGGAUAUAAGGAAUUCCAGCUCAUCCUUUGUUUCGUUCGAGUUUAAAGGAACGUCUGAGACAAGAGUAUUGCUGCCGUCAACUAACGUUGAGGCCACAGUG ((((((.....)))))).((((((....((....))......(((((((.(((....)))..)))))))..))))))........(((((..(((.(((((....))))))))..))))) ( -33.30) >DroEre_CAF1 51438 120 - 1 GUUAUUUGCUGGAUAACAUCGGAUAUGAGGAAUUCCAGCUCAUCCUUCGUCUCGUUCGAAUUUAAAGGAACAUCUGAGACAAGAGAAUUGCUGCCGUCAACUAACGUUGAGGCCACAGUG .......((((((.....(((....))).....))))))...(((((.(((((((((..........))))....)))))))).))((((..(((.(((((....))))))))..)))). ( -35.20) >DroYak_CAF1 56474 120 - 1 GUUAUUUGCUGAAUAACAUCGGAUAUAAGGAAUUCCAGCUCAUCCUUUGUCUCGUUCGAAUUUAAAGGAACAUCUGAGACAAGAUUAUUACUGCCGUCAACUAACGUUGAGGCCACAGUG .......(((((((((....((((....((....)).....))))((((((((((((..........))))....)))))))).)))))...(((.(((((....))))))))..)))). ( -30.40) >consensus GUUAUUUGCUGGAUAACAUCGGAUAUAAGGAAUUCCAGCUCAUCCUUUGUCUCGUUCGAAUUUAAAGGAACAUCUGAGACAAGAGUAUUGCUGCCGUCAACUAACGUUGAGGCCACAGUG ((((((.....)))))).((((((....((....))......(((((((..((....))...)))))))..)))))).........((((..(((.(((((....))))))))..)))). (-30.24 = -30.30 + 0.06)

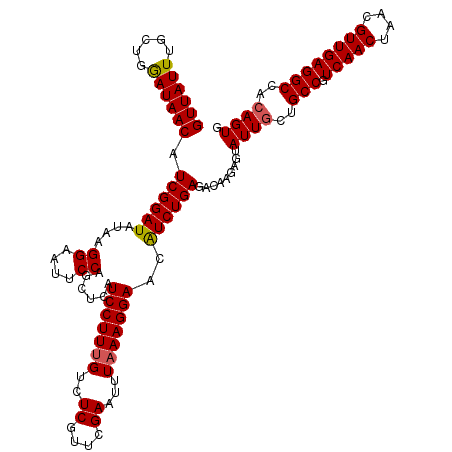
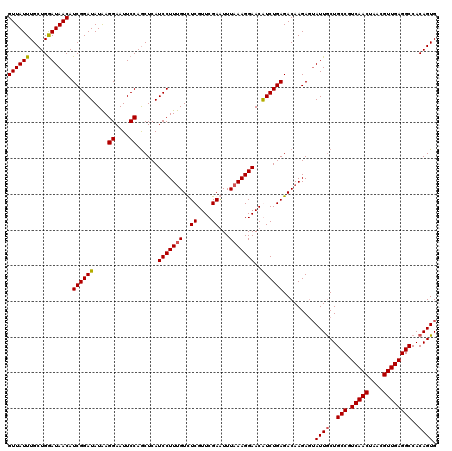
| Location | 1,434,978 – 1,435,098 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 90.97 |
| Mean single sequence MFE | -37.45 |
| Consensus MFE | -29.90 |
| Energy contribution | -29.52 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.619050 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1434978 120 - 20766785 UCAGUUGGGCGAAACUGGUCAGGCGAUUGGAAUCGUCGGCUGGUUGGGCGUUUGGCGGGAGCUUGUUUUGAUCGCCUUGGGUUAUUUGCUGGAUAACAUCGGAUAUAAGGAAUUCCAACU ...((..((((.(((..(((.((((((....)))))))))..)))...))))..))((((.(((((((((((....((.(((.....))).))....)))))).)))))...)))).... ( -38.80) >DroSec_CAF1 52157 120 - 1 UCAGUUGGGCGAAACUGGUCAGGCGAUUGGAAUCGUCGGCUGGUUGGGCGUUUGGCGGGAGCUUGUUUUGAUCGCCUUGGGUUAUUUGCUGGAUAACAUCGGAUAUAAGGAAUUCCAGCU ...((..((((.(((..(((.((((((....)))))))))..)))...))))..))((((.(((((((((((....((.(((.....))).))....)))))).)))))...)))).... ( -38.80) >DroEre_CAF1 51518 120 - 1 UCAGUUGGGCGAAAUUGGUCAGGCGAUUGGAAUCGUCGGCUGGUUGGGCUUUUGGCGGGAUCUUGUUUUGAUCGAUUUGAGUUAUUUGCUGGAUAACAUCGGAUAUGAGGAAUUCCAGCU (((((..(((..(((..(((.((((((....)))))))))..)))..))..)..))..((((.......))))....))).......((((((.....(((....))).....)))))). ( -35.60) >DroYak_CAF1 56554 114 - 1 UUAGUUGGGCGAAAUUGGUCAGGCGAUUGGAAUCGUCUGCUGGUUUGGCUUUUGUCGGG------UUUUCAUCGUUUUGAGUUAUUUGCUGAAUAACAUCGGAUAUAAGGAAUUCCAGCU ..((((((((.((((..(.((((((((....)))))))))..)))).))...((((.((------(..(((......)))(((((((...)))))))))).)))).........)))))) ( -36.60) >consensus UCAGUUGGGCGAAACUGGUCAGGCGAUUGGAAUCGUCGGCUGGUUGGGCGUUUGGCGGGAGCUUGUUUUGAUCGCCUUGAGUUAUUUGCUGGAUAACAUCGGAUAUAAGGAAUUCCAGCU ..((((((((..(((..(((.((((((....)))))))))..)))..))....(((((.............)))))((((((((((.....)))))).))))............)))))) (-29.90 = -29.52 + -0.38)

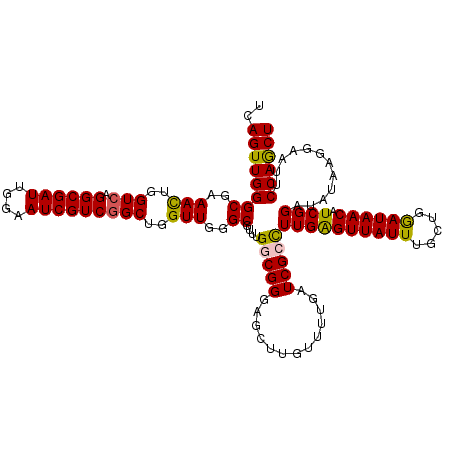
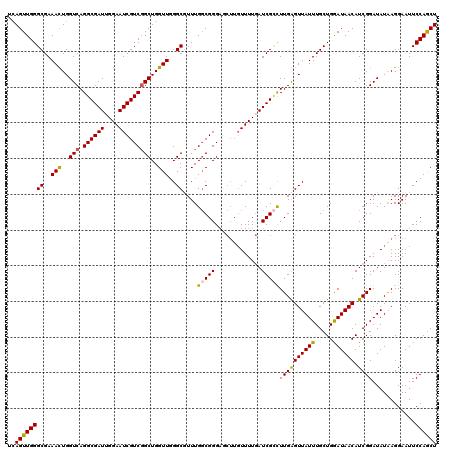
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:19 2006