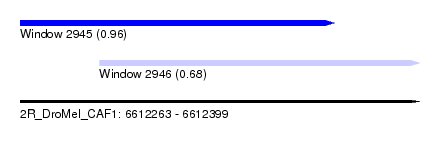
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,612,263 – 6,612,399 |
| Length | 136 |
| Max. P | 0.961463 |

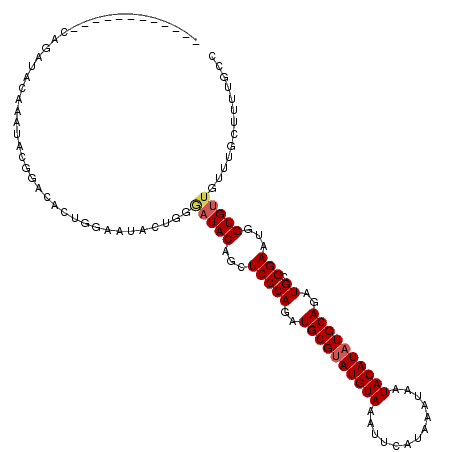
| Location | 6,612,263 – 6,612,370 |
|---|---|
| Length | 107 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 78.93 |
| Mean single sequence MFE | -27.32 |
| Consensus MFE | -19.84 |
| Energy contribution | -20.32 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.73 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961463 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6612263 107 + 20766785 ------------CAGAUACAAGUACGUAUAUGGGAAUGCUGGAAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAAUGGUGUUGUUUGCUUUUGCC ------------.........((((((((.(((((..((((.....))))))))).))))))))(((((....((((((((((...(((....))).....))))))))))..))))). ( -35.20) >DroSec_CAF1 164537 105 + 1 --------------GAUACAAAUACGGACACUGGAAUACUGGGAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAAUGGUGUUGUUUGCUUUUGCC --------------....((((..(((((.(..(....)..)(((((...(((((..((((((((((.............))))))))))..)).)))...))))))))))..)))).. ( -29.72) >DroSim_CAF1 151387 107 + 1 ------------CAGAUACAAAUACGGACACUGGAAUACUGGGAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAAUGGUGNNNNNNNNNNNNNNN ------------................((((......((((((......)))))).((((((((((.............))))))))))..........))))............... ( -22.82) >DroEre_CAF1 163494 119 + 1 CAGAAACCGAAACAGAUACAAAUACGAAUACUGAGAUACUGAGAUACAGCUCCCAGAUGCGCAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAUUGGUGUUGUUUGCUUUUGCC ............((((..((((((...((((..(....(((.....))).(((((..((((.(((((.............))))).))))..)).))))..))))))))))..)))).. ( -23.52) >DroYak_CAF1 171416 93 + 1 UAG------------AUACAA--------------GUACUGGGAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAUUGGUGUUGUUUGCUUUUGCC ...------------......--------------(((((((((......)))))).)))(((((((.............)))))))(((((.((((((.......)))))).))))). ( -25.32) >consensus ____________CAGAUACAAAUACGGACACUGGAAUACUGGGAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAAUGGUGUUGUUUGCUUUUGCC ..........................................(((((...(((((..((((((((((.............))))))))))..)).)))...)))))............. (-19.84 = -20.32 + 0.48)

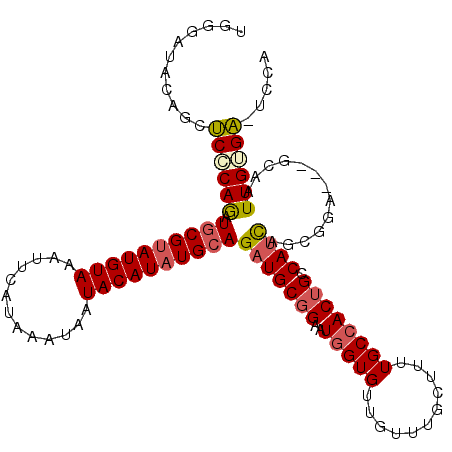
| Location | 6,612,290 – 6,612,399 |
|---|---|
| Length | 109 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 84.72 |
| Mean single sequence MFE | -31.28 |
| Consensus MFE | -19.20 |
| Energy contribution | -19.80 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681881 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6612290 109 + 20766785 UGGAAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAAUGGUGUUGUUUGCUUUUGCCACUGCCAUCAGCGGA---GCAAUUGUGA-UCCA .(((.((((((((((...((((((((((.............))))))))))(((((((..(((((..(....)...)))))))).)))).).)))---))...)))).-))). ( -35.62) >DroSec_CAF1 164562 109 + 1 UGGGAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAAUGGUGUUGUUUGCUUUUGCCACUGCCAUCAGCGGA---GCAAUUGUGA-UCCA ..(((((((((((((...((((((((((.............))))))))))(((((((..(((((..(....)...)))))))).)))).).)))---))...))).)-))). ( -34.92) >DroEre_CAF1 163533 109 + 1 UGAGAUACAGCUCCCAGAUGCGCAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAUUGGUGUUGUUUGCUUUUGCCACUGCCAUCAGCGAA---GCAAUUGGGA-UCCA ...........(((((((((.(((.(((((....((((((((((...(((....))).....))))))))))..)))))...))))))).((...---))...)))))-.... ( -27.40) >DroYak_CAF1 171429 109 + 1 UGGGAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAUUGGUGUUGUUUGCUUUUGCCACUGCCAUCAGCGGA---GCAAUUGGGA-UCCA ..((((.((((((((...((((((((((.............))))))))))(((((((..(((((..(....)...)))))))).)))).).)))---))...))..)-))). ( -34.02) >DroAna_CAF1 178504 99 + 1 -----------CCACAAAUACGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCCGCAUCGUG---UUUGUUUUUGCCACUGCCACUGAGGCACCCACCAUUGUGGCCCCA -----------..(((((((((((((((.............)))))))...((((...)))))))---)))))....(((((((((.....)))).........))))).... ( -24.42) >consensus UGGGAUACAGCUCCCAGAUGCGUAUGUAAAUUCAUAAAUAAUACAUAUGCAGAUGCGGAAUGGUGUUGUUUGCUUUUGCCACUGCCAUCAGCGGA___GCAAUUGUGA_UCCA ...........((((((.((((((((((.............))))))))))(((((((..(((((...........)))))))).)))).............))))))..... (-19.20 = -19.80 + 0.60)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:45 2006