
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,609,032 – 6,609,136 |
| Length | 104 |
| Max. P | 0.862929 |

| Location | 6,609,032 – 6,609,136 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.30 |
| Mean single sequence MFE | -21.74 |
| Consensus MFE | -15.72 |
| Energy contribution | -16.08 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.862929 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6609032 104 - 20766785 UACAAUUAUGUAUGCUUCGGCUGCUGCCUGAUAAAUUUGUAUUUAUUAGACUCAUACAUCAAGUUGGCAUCU---UUGUGGGUCUCUCUCCGUCUAUAUCCCA-------------CUCG ..((((((((((((..(((((....)))((((((((....))))))))))..)))))))..)))))......---..(((((.................))))-------------)... ( -25.33) >DroSim_CAF1 148188 104 - 1 UACAAUUAUGUAUGCUUCGGCUGCUGCCUGAUAAAUUUGUAUUUAUUAGACUCAUACAUCAAGUUGGCAUCU---UUGUGGGUCUCUCUCCGUCUAUAUCCCA-------------CUCG ..((((((((((((..(((((....)))((((((((....))))))))))..)))))))..)))))......---..(((((.................))))-------------)... ( -25.33) >DroEre_CAF1 160433 95 - 1 UACAAUUAUGUAUGCUUCGGCUGCUGCCUGAUAAAUUUGUAUUUAUUAGACUCAUACAUCAAGUUGGCAUCU---UUGUGGGUC----UCUGUCUAUAUCCC------------------ ..((((((((((((..(((((....)))((((((((....))))))))))..)))))))..)))))......---.((((((..----....))))))....------------------ ( -21.10) >DroWil_CAF1 201269 84 - 1 UACAAUUAUAUAUGCUUCGGUUAGUGGCUGAUAAAUUUGUAUUUAUUAGACUCAUACAUCAAGUUGGCAUCUCCUUUGUGAAUG------------------------------------ (((((......(((((..(((..(((((((((((((....))))))))).).)))..))).....))))).....)))))....------------------------------------ ( -14.70) >DroYak_CAF1 168182 113 - 1 UACAAUUAUGUAUGCUUCGGCUGCUGCCUGAUAAAUUUGUAUUUAUUAGACUCAUACAUCAAGUUGGCAUCU---UUGUGGCUC----UCCGUCUAUAUCCCAUACCCAUAUCCCCCUCG ..((((((((((((..(((((....)))((((((((....))))))))))..)))))))..)))))......---.(((((...----............)))))............... ( -20.56) >DroAna_CAF1 175202 108 - 1 UACAAUUAUGUAUGCUUCGGCUGCUGCCUGAUAAAUUUGUAUUUAUUAGACUCAUACUUCAAGUUGCCAUCU---UUGUGGGUCUCCCCCGGUGGAUCUCUCUUAUCC-UAU-------- ..(((((..(((((..(((((....)))((((((((....))))))))))..)))))....)))))(((((.---..(.(((....))))))))).............-...-------- ( -23.40) >consensus UACAAUUAUGUAUGCUUCGGCUGCUGCCUGAUAAAUUUGUAUUUAUUAGACUCAUACAUCAAGUUGGCAUCU___UUGUGGGUC____UCCGUCUAUAUCCCA_________________ ..((((((((((((..(((((....)))((((((((....))))))))))..)))))))..)))))...................................................... (-15.72 = -16.08 + 0.36)

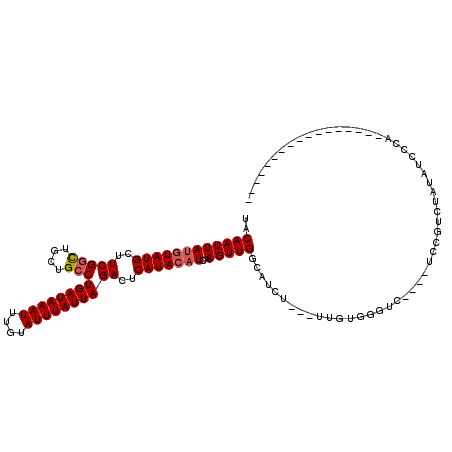
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:42 2006