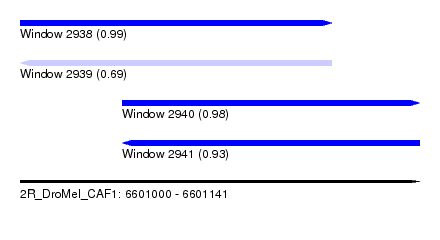
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,601,000 – 6,601,141 |
| Length | 141 |
| Max. P | 0.991788 |

| Location | 6,601,000 – 6,601,110 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -42.65 |
| Consensus MFE | -35.40 |
| Energy contribution | -36.03 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.97 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.991788 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
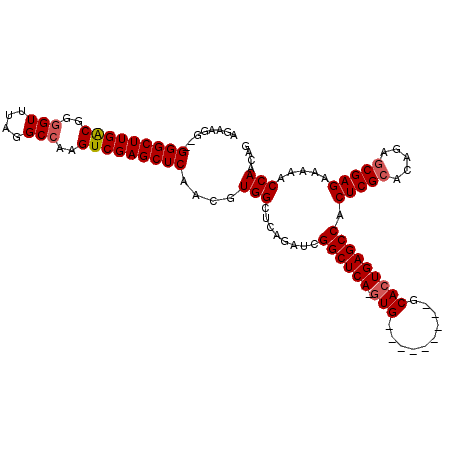
>2R_DroMel_CAF1 6601000 110 + 20766785 AGAAGG--GGGCUUGACGGUGUUUAGGCCAAGUCGAGCUCAACGUGGCUCAGAUCGGCUCAAGUGAGCCCCAAUGCACUGAGCCACUCGCACAGAGCGAGAAAAACCAACAG ....((--((((((((((((......)))..)))))))))...(((((((((...(((((....)))))........)))))))))((((.....))))......))..... ( -46.00) >DroSec_CAF1 153484 100 + 1 AGAAGG--GGGCUUGGCGGGGUUUAGGCCAAGUCGAGCUCAACGUGGCUCAGAUCGGCUCA-GUG---------GCACUGAGCCACUCGCUCAGGACGAGAAAAACCAACAG ....((--(((((((((..(((....)))..)))))))))...(((((((((....(((..-..)---------)).)))))))))...(((.....))).....))..... ( -41.60) >DroSim_CAF1 140281 102 + 1 AGAAGGGGGGGCUUGGCGGGGUUUAGGCCAAGUCGAGCUCAACGUGGCUCAGAUCGGCUCA-GUG---------GCACUGAGCCACUCGCACAGAGCGAGAAAAACCAACGG ........(((((((((..(((....)))..)))))))))..(((((........((((((-(..---------...))))))).(((((.....))))).....)).))). ( -44.80) >DroYak_CAF1 159979 95 + 1 AGAAGG--GGGCUUGACGCGGUUUGGGCCAAGUCGAGCUCAACGUGGCUCCGAUCGGCUCA-G--------------GUGAGCCACUCGCACAGAGCGAGAAAAACCAACAG ....((--(((((((((..(((....)))..)))))))))...(((((((..(((......-)--------------)))))))))((((.....))))......))..... ( -38.20) >consensus AGAAGG__GGGCUUGACGGGGUUUAGGCCAAGUCGAGCUCAACGUGGCUCAGAUCGGCUCA_GUG_________GCACUGAGCCACUCGCACAGAGCGAGAAAAACCAACAG ........(((((((((..(((....)))..)))))))))....(((........((((((.(((..........))))))))).(((((.....))))).....))).... (-35.40 = -36.03 + 0.62)

| Location | 6,601,000 – 6,601,110 |
|---|---|
| Length | 110 |
| Sequences | 4 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 87.11 |
| Mean single sequence MFE | -35.40 |
| Consensus MFE | -29.81 |
| Energy contribution | -30.00 |
| Covariance contribution | 0.19 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.84 |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.687272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6601000 110 - 20766785 CUGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAGUGCAUUGGGGCUCACUUGAGCCGAUCUGAGCCACGUUGAGCUCGACUUGGCCUAAACACCGUCAAGCCC--CCUUCU ..((((((((..(((((.....))))(((((((((........(((((....)))))...))))))))))..)))).))))(((((.........)))))....--...... ( -38.10) >DroSec_CAF1 153484 100 - 1 CUGUUGGUUUUUCUCGUCCUGAGCGAGUGGCUCAGUGC---------CAC-UGAGCCGAUCUGAGCCACGUUGAGCUCGACUUGGCCUAAACCCCGCCAAGCCC--CCUUCU ...(((((....(((((.....)))))((((((((...---------..)-)))))))......((((.((((....)))).)))).........)))))....--...... ( -32.60) >DroSim_CAF1 140281 102 - 1 CCGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAGUGC---------CAC-UGAGCCGAUCUGAGCCACGUUGAGCUCGACUUGGCCUAAACCCCGCCAAGCCCCCCCUUCU ...(((((....(((((.....)))))((((((((...---------..)-)))))))......((((.((((....)))).)))).........)))))............ ( -35.30) >DroYak_CAF1 159979 95 - 1 CUGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAC--------------C-UGAGCCGAUCGGAGCCACGUUGAGCUCGACUUGGCCCAAACCGCGUCAAGCCC--CCUUCU .....(((((..(((((.....)))))((((((..--------------.-.))))))...((.((((.((((....)))).)))))))))))((.....))..--...... ( -35.60) >consensus CUGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAGUGC_________CAC_UGAGCCGAUCUGAGCCACGUUGAGCUCGACUUGGCCUAAACCCCGCCAAGCCC__CCUUCU .....(((((..(((((.....)))))((((((((((..........))).)))))))......((((.((((....)))).))))............)))))......... (-29.81 = -30.00 + 0.19)

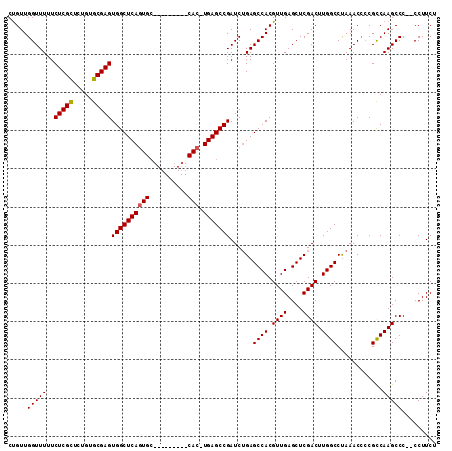
| Location | 6,601,036 – 6,601,141 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 84.46 |
| Mean single sequence MFE | -35.75 |
| Consensus MFE | -25.31 |
| Energy contribution | -26.50 |
| Covariance contribution | 1.19 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.94 |
| Structure conservation index | 0.71 |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.983766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
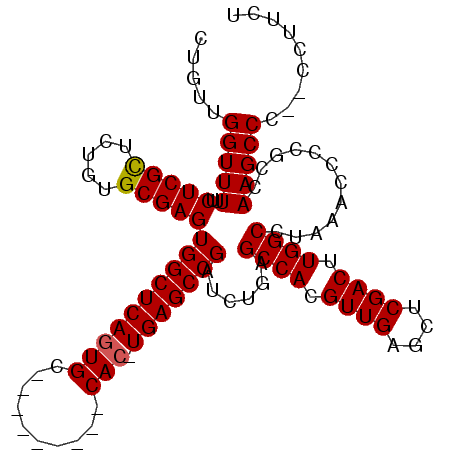
>2R_DroMel_CAF1 6601036 105 + 20766785 UCAACGUGGCUCAGAUCGGCUCAAGUGAGCCCCAAUGCACUGAGCCACUCGCACAGAGCGAGAAAAACCAACAGAGACCA-GAGACUGUUCAAGCGUCUCUGGAGC .....(((((((((...(((((....)))))........)))))))))((((.....))))................(((-(((((.((....))))))))))... ( -42.30) >DroSec_CAF1 153520 96 + 1 UCAACGUGGCUCAGAUCGGCUCA-GUG---------GCACUGAGCCACUCGCUCAGGACGAGAAAAACCAACAGAGACCAACAGACUGUUCAAGCGUCUGUGGACC ......((((((.....((((((-(..---------...))))))).(((((....).))))...........))).)))((((((.((....))))))))..... ( -32.90) >DroSim_CAF1 140319 96 + 1 UCAACGUGGCUCAGAUCGGCUCA-GUG---------GCACUGAGCCACUCGCACAGAGCGAGAAAAACCAACGGAGACCAACAGACUGUUCAAGCGUCUGUGGAGC ((..(((((........((((((-(..---------...))))))).(((((.....))))).....)).)))..))(((.(((((.((....))))))))))... ( -37.00) >DroYak_CAF1 160015 86 + 1 UCAACGUGGCUCCGAUCGGCUCA-G--------------GUGAGCCACUCGCACAGAGCGAGAAAAACCAACAGAAACCG-----CUGUUCAAGCGUCUCUGGAGC ........(((((....(((((.-.--------------..))))).(((((.....)))))..........(((...((-----((.....))))..)))))))) ( -30.80) >consensus UCAACGUGGCUCAGAUCGGCUCA_GUG_________GCACUGAGCCACUCGCACAGAGCGAGAAAAACCAACAGAGACCA_CAGACUGUUCAAGCGUCUCUGGAGC ((....(((........((((((.(((..........))))))))).(((((.....))))).....)))...))..(((.(((((.((....))))))))))... (-25.31 = -26.50 + 1.19)

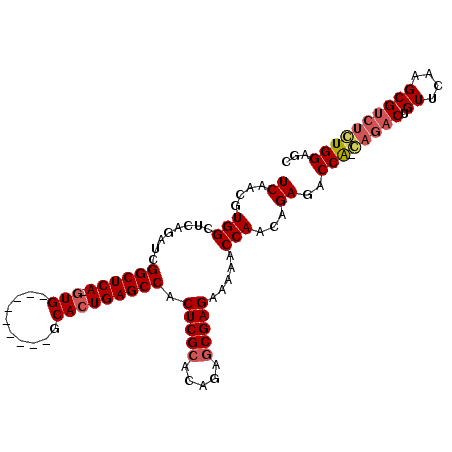
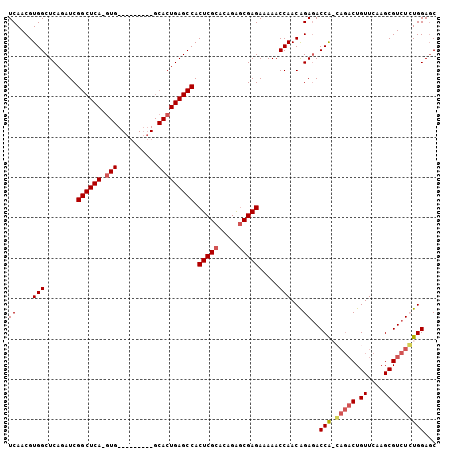
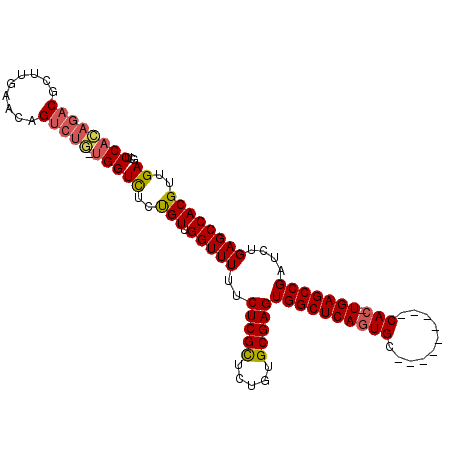
| Location | 6,601,036 – 6,601,141 |
|---|---|
| Length | 105 |
| Sequences | 4 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 84.46 |
| Mean single sequence MFE | -37.47 |
| Consensus MFE | -28.01 |
| Energy contribution | -28.82 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.23 |
| Structure conservation index | 0.75 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931235 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6601036 105 - 20766785 GCUCCAGAGACGCUUGAACAGUCUC-UGGUCUCUGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAGUGCAUUGGGGCUCACUUGAGCCGAUCUGAGCCACGUUGA ...((((((((.........)))))-)))................((((.....))))(((((((((........(((((....)))))...)))))))))..... ( -42.70) >DroSec_CAF1 153520 96 - 1 GGUCCACAGACGCUUGAACAGUCUGUUGGUCUCUGUUGGUUUUUCUCGUCCUGAGCGAGUGGCUCAGUGC---------CAC-UGAGCCGAUCUGAGCCACGUUGA (..(.((((((.........)))))).)..).....((((((..(((((.....)))))((((((((...---------..)-)))))))....))))))...... ( -34.50) >DroSim_CAF1 140319 96 - 1 GCUCCACAGACGCUUGAACAGUCUGUUGGUCUCCGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAGUGC---------CAC-UGAGCCGAUCUGAGCCACGUUGA ...((((((((.........))))).)))((..(((.(((((..(((((.....)))))((((((((...---------..)-)))))))....))))))))..)) ( -37.90) >DroYak_CAF1 160015 86 - 1 GCUCCAGAGACGCUUGAACAG-----CGGUUUCUGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAC--------------C-UGAGCCGAUCGGAGCCACGUUGA ((((((((((((((.....))-----).))))))...((((...(((((.....))))).(((((..--------------.-.))))))))))))))........ ( -34.80) >consensus GCUCCACAGACGCUUGAACAGUCUG_UGGUCUCUGUUGGUUUUUCUCGCUCUGUGCGAGUGGCUCAGUGC_________CAC_UGAGCCGAUCUGAGCCACGUUGA ...((((((((.........))))).)))((..(((.(((((..(((((.....)))))((((((((((..........))).)))))))....))))))))..)) (-28.01 = -28.82 + 0.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:40 2006