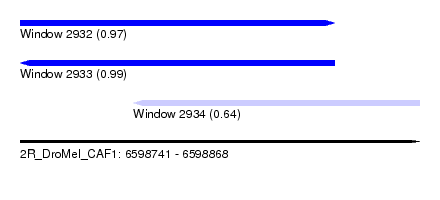
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,598,741 – 6,598,868 |
| Length | 127 |
| Max. P | 0.987359 |

| Location | 6,598,741 – 6,598,841 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -29.30 |
| Consensus MFE | -23.77 |
| Energy contribution | -24.53 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.04 |
| Mean z-score | -3.11 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974639 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
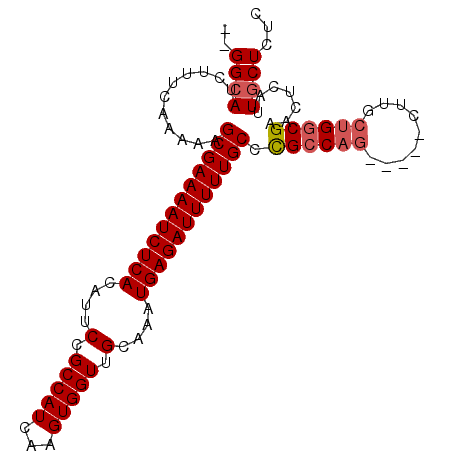
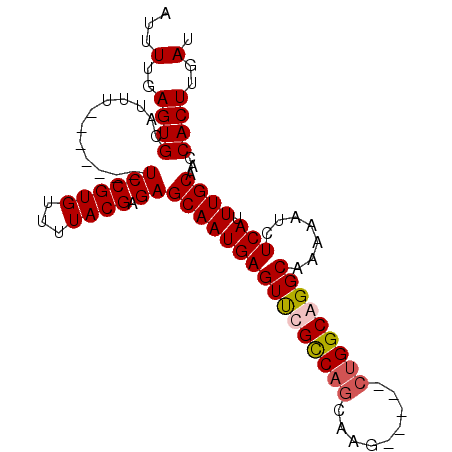
>2R_DroMel_CAF1 6598741 100 + 20766785 --GGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCUGCCACCACCUCUCGCUGGCGAACUCAUUGCUCUC --((((...(((.....((((((((((((....(.(((((...))))).)....))))))))))))..((((.(.......).))))))).....))))... ( -27.80) >DroSec_CAF1 151236 95 + 1 --GGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCCGCCAG-----CUUGCUGGCGAACUCAUUGCUCUC --((((...........((((((((((((....(.(((((...))))).)....)))))))))))).((((((-----....)))))).......))))... ( -33.30) >DroSim_CAF1 138032 95 + 1 --GGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCCGCCAG-----CUUGCUGGCGAACUCAUUGCUCUC --((((...........((((((((((((....(.(((((...))))).)....)))))))))))).((((((-----....)))))).......))))... ( -33.30) >DroYak_CAF1 157646 93 + 1 GUGGAAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCUGCCAG---------CGGACGGACUCAUUGCUCUC ...((.(((........((((((((((((....(.(((((...))))).)....))))))))))))(((....---------)))..))).))......... ( -22.80) >consensus __GGCAUCUUUCAAAAAGCGAAAAUCUCACAUUCCGCCAUCAAGUGGUUGCAAAUGAGAUUUUUGCCCGCCAG_____CUUGCUGGCGAACUCAUUGCUCUC ..((((...........((((((((((((....(.(((((...))))).)....)))))))))))).((((((.........)))))).......))))... (-23.77 = -24.53 + 0.75)


| Location | 6,598,741 – 6,598,841 |
|---|---|
| Length | 100 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 90.86 |
| Mean single sequence MFE | -32.32 |
| Consensus MFE | -28.58 |
| Energy contribution | -29.20 |
| Covariance contribution | 0.62 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.88 |
| SVM decision value | 2.08 |
| SVM RNA-class probability | 0.987359 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6598741 100 - 20766785 GAGAGCAAUGAGUUCGCCAGCGAGAGGUGGUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCC-- ..((((.....))))((((.(....).))))(((((((.((((((((((...(..(((.....)))..)...)))))))))).)))..((....))))))-- ( -33.10) >DroSec_CAF1 151236 95 - 1 GAGAGCAAUGAGUUCGCCAGCAAG-----CUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCC-- ((((((.....(((((((((....-----))))))))).((((((((((...(..(((.....)))..)...)))))))))).))))))...........-- ( -36.00) >DroSim_CAF1 138032 95 - 1 GAGAGCAAUGAGUUCGCCAGCAAG-----CUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCC-- ((((((.....(((((((((....-----))))))))).((((((((((...(..(((.....)))..)...)))))))))).))))))...........-- ( -36.00) >DroYak_CAF1 157646 93 - 1 GAGAGCAAUGAGUCCGUCCG---------CUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUUCCAC ((((((.....(((.(((..---------..))).))).((((((((((...(..(((.....)))..)...)))))))))).))))))............. ( -24.20) >consensus GAGAGCAAUGAGUUCGCCAGCAAG_____CUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAUGGCGGAAUGUGAGAUUUUCGCUUUUUGAAAGAUGCC__ ((((((.....(((((((((.........))))))))).((((((((((...(..(((.....)))..)...)))))))))).))))))............. (-28.58 = -29.20 + 0.62)

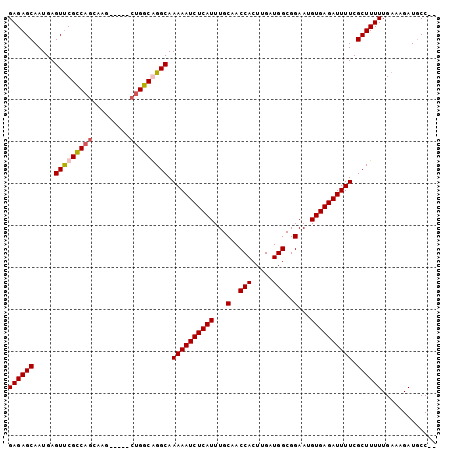
| Location | 6,598,777 – 6,598,868 |
|---|---|
| Length | 91 |
| Sequences | 4 |
| Columns | 98 |
| Reading direction | reverse |
| Mean pairwise identity | 87.50 |
| Mean single sequence MFE | -24.48 |
| Consensus MFE | -19.71 |
| Energy contribution | -20.34 |
| Covariance contribution | 0.63 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642468 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
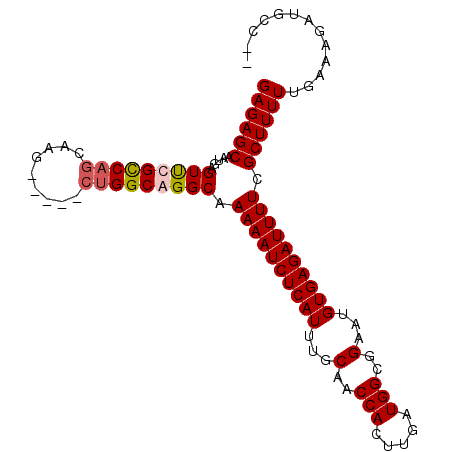
>2R_DroMel_CAF1 6598777 91 - 20766785 AUUUUGAGUGCAUUU-------UCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGCGAGAGGUGGUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAU ...(..((((.....-------((((((...)))).))((((((((..(((((.(....).)))))(....)......)))).))))...))))..). ( -24.60) >DroSec_CAF1 151272 86 - 1 AUUUUGAGUGAAUUU-------UCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGCAAG-----CUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAU ...(..((((.....-------((((((...)))).))((((((((((((((((....-----))))))))).......))).))))...))))..). ( -27.81) >DroSim_CAF1 138068 86 - 1 AUUUUGAGUGCAUUU-------UCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGCAAG-----CUGGCGGGCAAAAAUCUCAUUUGCAACCACUUGAU ...(..((((.....-------((((((...)))).))((((((((((((((((....-----))))))))).......))).))))...))))..). ( -27.11) >DroYak_CAF1 157684 89 - 1 AUUUUGAGUGCAUUUUACGUGUUCCGUGUUUUACGAGAGCAAUGAGUCCGUCCG---------CUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAU ...(..((((..........((((((((...)))).))))((((((.......(---------((....)))......))))))......))))..). ( -18.42) >consensus AUUUUGAGUGCAUUU_______UCCGUGUUUUACGAGAGCAAUGAGUUCGCCAGCAAG_____CUGGCAGGCAAAAAUCUCAUUUGCAACCACUUGAU ...(..((((............((((((...)))).))((((((((((((((((.........))))))))).......))).))))...))))..). (-19.71 = -20.34 + 0.63)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:33 2006