
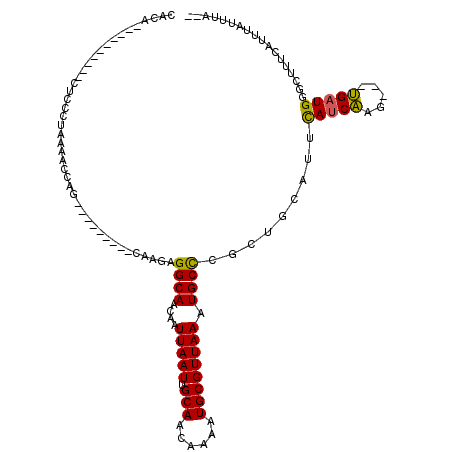
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,598,603 – 6,598,707 |
| Length | 104 |
| Max. P | 0.633836 |

| Location | 6,598,603 – 6,598,707 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 74.88 |
| Mean single sequence MFE | -21.15 |
| Consensus MFE | -11.19 |
| Energy contribution | -10.80 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.53 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633836 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6598603 104 - 20766785 CACACCAC--CACACUACCUAAAACCAG---------CAAGAGGCAACAAUUAAUUGCAACAAAAUGCGUUAAAUGCCCGAUGCAUUCAUCAAG---UGAUGGGUUUUCAUUUAUUUA-- ........--..........((((((.(---------((.(.((((....(((((.(((......)))))))).)))))..)))...((((...---.))))))))))..........-- ( -20.00) >DroSec_CAF1 151094 106 - 1 CACACCACAACACACUCCCUAAGACCAG---------CAAGCGGCAACUAUUAAUUGCAACAAAAUGCGUUAAAUGCUCGCCGCAUUCAUCAAG---UGAUGGGCUUUCAUUUAUUUA-- ......................((..((---------(..(((((...((((...((((......))))...))))...)))))..(((((...---.)))))))).)).........-- ( -20.80) >DroSim_CAF1 137890 106 - 1 CACACCACAACACACUCCCUAAGACCAG---------CAAGCGGCAACUAUUAAUUGCAACAAAAUGCGUUAAAUGCCCGCCGCAUUCAUCAAG---UGAUGGGCUUUCAUUUAUUUA-- ......................((..((---------(..(((((...((((...((((......))))...))))...)))))..(((((...---.)))))))).)).........-- ( -20.80) >DroEre_CAF1 150327 96 - 1 CGCA----------CACCCCAAAAUCGG---------CAUGAGGCAAAAAUUAAUUGCAACAAAAUGCGUUAAAUGCCUGCUGCAUGCAUCAUG---UGUUGGGCCUUCGUUUAUUUA-- ....----------...(((((.....(---------(((((((((....(((((.(((......)))))))).))))(((.....))))))))---).)))))..............-- ( -22.30) >DroYak_CAF1 157498 105 - 1 CACA----------CUCCCUAGAACCUAGAAGCCGAAAAAGAGGCAAGAAUUAAUUGCAACAAAAUGCGUUAAAUGCCGGCUGCAUUCAUCAAG---UGAUGGCCUUUCAUUUAUUUA-- ....----------....((((...)))).....((((....((((....(((((.(((......)))))))).))))(((((((((.....))---)).))))))))).........-- ( -19.10) >DroAna_CAF1 164546 100 - 1 CACA----------CUCACUAAAAUUCG---------AAAGAGGCAGAAAUUAAUUGCAACAAAAUGCGUUAAAUGCCUUUUACAUUUAUCCAAAUUGGAUGGGCAUAGA-GGAUGAAUG ((..----------(((..........(---------.((((((((....(((((.(((......)))))))).)))))))).).(((((((.....)))))))....))-)..)).... ( -23.90) >consensus CACA__________CUCCCUAAAACCAG_________CAAGAGGCAACAAUUAAUUGCAACAAAAUGCGUUAAAUGCCCGCUGCAUUCAUCAAG___UGAUGGGCUUUCAUUUAUUUA__ ..........................................((((....(((((.(((......)))))))).)))).........(((((.....))))).................. (-11.19 = -10.80 + -0.39)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:31 2006