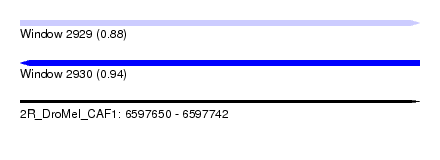
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,597,650 – 6,597,742 |
| Length | 92 |
| Max. P | 0.938670 |

| Location | 6,597,650 – 6,597,742 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -13.98 |
| Consensus MFE | -10.23 |
| Energy contribution | -10.53 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878574 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
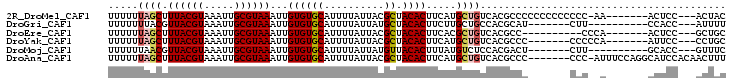
>2R_DroMel_CAF1 6597650 92 + 20766785 UUUUUUAGCUUUACGUAAAUUGCGUAAAUUGUGUGCAUUUUAUUACGCUACACUUCAUGCUGUCACGCCCCCCCCCCCCC-AA-------ACUCC---ACUAC .....(((((((((((.....)))))))..((((((..........)).)))).....))))..................-..-------.....---..... ( -13.10) >DroGri_CAF1 107154 83 + 1 UUUUUUUACGUUACGUAAAUUGCGUAAAUUGUGUGCAUUUUAUUAUGCUACACUUCUUGCUGCCACGCAU-------CUU----------CCACC---AUUUU ....(((((((..........)))))))..((((((((......)))).))))....(((......))).-------...----------.....---..... ( -14.60) >DroEre_CAF1 149403 83 + 1 UUUUUUAGCUUUACGUAAAUUGCGUAAAUUGUGUGCAUUUUAUUACGCUACACUUCACGCUGUCACGCC----------CCCA-------ACUCC---GCUGC .....(((((((((((.....)))))))..((((((..........)).)))).....)))).......----------....-------.....---..... ( -13.40) >DroYak_CAF1 156552 86 + 1 UUUUUUAGCUUUACGUAAAUUGCGUAAAUUGUGUGCAUUUUAUUACGCUACACUUCAUGCUGUCACGCCC-------CCCCCA-------AUUCC---CCUGC .....(((((((((((.....)))))))..((((((..........)).)))).....))))........-------......-------.....---..... ( -13.10) >DroMoj_CAF1 117160 83 + 1 UUUUUUAACGUUACGUAAAUUGCGUAAAUUGUGUGCAUUUUAUUAUGUUACACUUUAUGUCUCCACGACU-------CUU----------GCACC---GUUUC ......((((((((((.....))))))...((((((((......))).))))).....(((.....))).-------...----------....)---))).. ( -13.50) >DroAna_CAF1 163409 95 + 1 UUUUUUAGCUUUACGUAAAUUGCGUAAAUUGUGUGCAUUUUAUUACGCUACACUUCAUGCUGUCACGCCC-------CCC-AUUUCCAGGCAUCCACAACUUU .....(((((((((((.....)))))))..((((((..........)).)))).....))))....(((.-------...-.......)))............ ( -16.20) >consensus UUUUUUAGCUUUACGUAAAUUGCGUAAAUUGUGUGCAUUUUAUUACGCUACACUUCAUGCUGUCACGCCC_______CCC__A_______ACUCC___ACUUC .....((((.((((((.....))))))...((((((..........)).)))).....))))......................................... (-10.23 = -10.53 + 0.31)
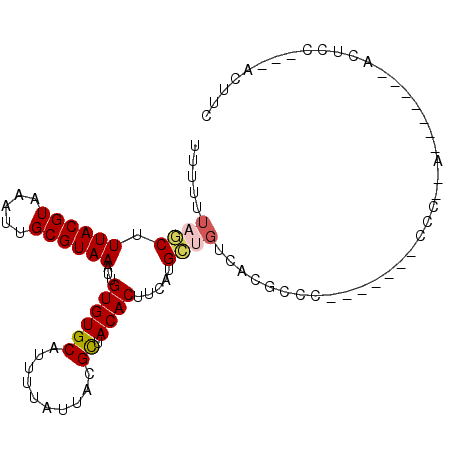
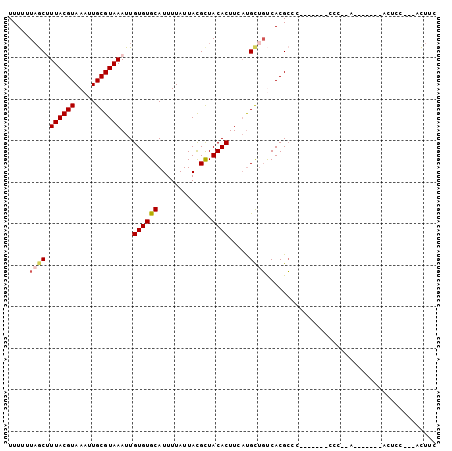
| Location | 6,597,650 – 6,597,742 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 78.13 |
| Mean single sequence MFE | -19.17 |
| Consensus MFE | -12.63 |
| Energy contribution | -12.72 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.66 |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.938670 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6597650 92 - 20766785 GUAGU---GGAGU-------UU-GGGGGGGGGGGGGCGUGACAGCAUGAAGUGUAGCGUAAUAAAAUGCACACAAUUUACGCAAUUUACGUAAAGCUAAAAAA .....---..(((-------((-...(.(..((..(((((((.....)..((((.((((......))))))))...))))))..))..).).)))))...... ( -19.10) >DroGri_CAF1 107154 83 - 1 AAAAU---GGUGG----------AAG-------AUGCGUGGCAGCAAGAAGUGUAGCAUAAUAAAAUGCACACAAUUUACGCAAUUUACGUAACGUAAAAAAA ....(---(((((----------(..-------.(((......)))....((((.((((......))))))))..))))).)).((((((...)))))).... ( -17.10) >DroEre_CAF1 149403 83 - 1 GCAGC---GGAGU-------UGGG----------GGCGUGACAGCGUGAAGUGUAGCGUAAUAAAAUGCACACAAUUUACGCAAUUUACGUAAAGCUAAAAAA .....---..(((-------(...----------.((((((..(((((((((((.((((......))))))))..)))))))...))))))..))))...... ( -24.00) >DroYak_CAF1 156552 86 - 1 GCAGG---GGAAU-------UGGGGG-------GGGCGUGACAGCAUGAAGUGUAGCGUAAUAAAAUGCACACAAUUUACGCAAUUUACGUAAAGCUAAAAAA (((.(---..(((-------((.(..-------((((......)).....((((.((((......))))))))..))..).)))))..).)...))....... ( -16.80) >DroMoj_CAF1 117160 83 - 1 GAAAC---GGUGC----------AAG-------AGUCGUGGAGACAUAAAGUGUAACAUAAUAAAAUGCACACAAUUUACGCAAUUUACGUAACGUUAAAAAA ..(((---(((((----------(..-------.((..((...((((...))))..))..))....))))).....(((((.......)))))))))...... ( -15.10) >DroAna_CAF1 163409 95 - 1 AAAGUUGUGGAUGCCUGGAAAU-GGG-------GGGCGUGACAGCAUGAAGUGUAGCGUAAUAAAAUGCACACAAUUUACGCAAUUUACGUAAAGCUAAAAAA ...((..((..(.(((......-)))-------.).))..))(((.....((((.((((......))))))))..((((((.......)))))))))...... ( -22.90) >consensus GAAGU___GGAGU_______U__GGG_______GGGCGUGACAGCAUGAAGUGUAGCGUAAUAAAAUGCACACAAUUUACGCAAUUUACGUAAAGCUAAAAAA ...................................((((((..((.((((((((.((((......))))))))..)))).))...))))))............ (-12.63 = -12.72 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:30 2006