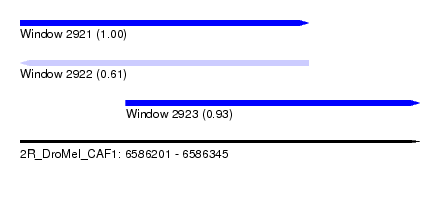
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,586,201 – 6,586,345 |
| Length | 144 |
| Max. P | 0.998288 |

| Location | 6,586,201 – 6,586,305 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -21.30 |
| Consensus MFE | -21.30 |
| Energy contribution | -21.30 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -3.30 |
| Structure conservation index | 1.00 |
| SVM decision value | 3.06 |
| SVM RNA-class probability | 0.998288 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
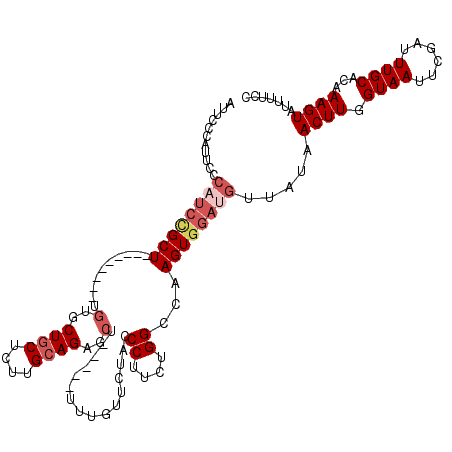
>2R_DroMel_CAF1 6586201 104 + 20766785 CCACUUCACUCCGCAUCGCCAUCCACAUCCCCUUUCGCAUUCCCAUUUCCAUCCGCUUGUGCUGCUUUUGCAGAGCUUUUGUUCUACCCUUCUGGCCAAGUGGA ...................................................((((((((.(((((....))(((((....)))))........))))))))))) ( -21.30) >DroSec_CAF1 138877 86 + 1 CCACUUCACUCCGCAUCCCC------------------AUUCCCAUUCCCAUCCGCUUGUGCUGCUCUUGCAGAGCUUUUGUUCUACCCUUCUGGCCAAGUGGA ....................------------------.............((((((((.(((((....))(((((....)))))........))))))))))) ( -21.30) >DroSim_CAF1 125178 86 + 1 CCACUUCACUCCGCAUCCCC------------------AUUCCCAUUCCCAUCCGCUUGUGCUGCUCUUGCAGAGCUUUUGUUCUACCCUUCUGGCCAAGUGGA ....................------------------.............((((((((.(((((....))(((((....)))))........))))))))))) ( -21.30) >consensus CCACUUCACUCCGCAUCCCC__________________AUUCCCAUUCCCAUCCGCUUGUGCUGCUCUUGCAGAGCUUUUGUUCUACCCUUCUGGCCAAGUGGA ...................................................((((((((.(((((....))(((((....)))))........))))))))))) (-21.30 = -21.30 + 0.00)

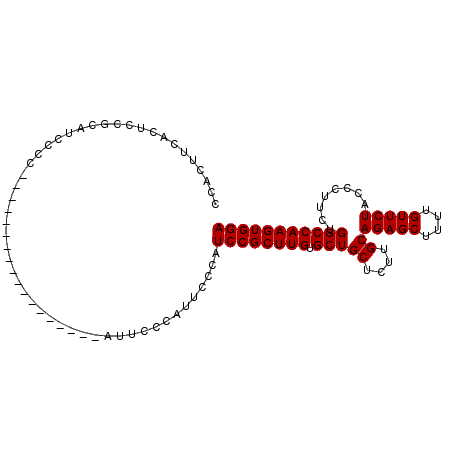
| Location | 6,586,201 – 6,586,305 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 85.71 |
| Mean single sequence MFE | -23.13 |
| Consensus MFE | -20.60 |
| Energy contribution | -20.60 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.35 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.608435 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6586201 104 - 20766785 UCCACUUGGCCAGAAGGGUAGAACAAAAGCUCUGCAAAAGCAGCACAAGCGGAUGGAAAUGGGAAUGCGAAAGGGGAUGUGGAUGGCGAUGCGGAGUGAAGUGG .((((((.(((.....))).........((((((((...((..((((.....((....)).....(.(....).)..))))....))..)))))))).)))))) ( -27.60) >DroSec_CAF1 138877 86 - 1 UCCACUUGGCCAGAAGGGUAGAACAAAAGCUCUGCAAGAGCAGCACAAGCGGAUGGGAAUGGGAAU------------------GGGGAUGCGGAGUGAAGUGG .((((((.(((.....))).........((((((((....((...((..(......)..))....)------------------)....)))))))).)))))) ( -20.90) >DroSim_CAF1 125178 86 - 1 UCCACUUGGCCAGAAGGGUAGAACAAAAGCUCUGCAAGAGCAGCACAAGCGGAUGGGAAUGGGAAU------------------GGGGAUGCGGAGUGAAGUGG .((((((.(((.....))).........((((((((....((...((..(......)..))....)------------------)....)))))))).)))))) ( -20.90) >consensus UCCACUUGGCCAGAAGGGUAGAACAAAAGCUCUGCAAGAGCAGCACAAGCGGAUGGGAAUGGGAAU__________________GGGGAUGCGGAGUGAAGUGG .((((((.(((.....))).........((((((((......((....))..((....)).............................)))))))).)))))) (-20.60 = -20.60 + -0.00)


| Location | 6,586,239 – 6,586,345 |
|---|---|
| Length | 106 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 83.03 |
| Mean single sequence MFE | -27.46 |
| Consensus MFE | -15.78 |
| Energy contribution | -17.26 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.57 |
| SVM decision value | 1.19 |
| SVM RNA-class probability | 0.928802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6586239 106 + 20766785 AUUCCCAUUUCCAUCCGCU---------UGUGCUGCUUUUGCAGAGCU-----UUUGUUCUACCCUUCUGGCCAAGUGGAUGUUAUAACUUGGUAAUUCGAUUUGCACAAAGUAUUUUCC ...........((((((((---------((.(((((....))(((((.-----...)))))........))))))))))))).....((((.((((......))))...))))....... ( -30.20) >DroSec_CAF1 138897 106 + 1 AUUCCCAUUCCCAUCCGCU---------UGUGCUGCUCUUGCAGAGCU-----UUUGUUCUACCCUUCUGGCCAAGUGGAUGUUAUAACUUGGUAAUUCGAUUUGCACAAAGUAUUUUCC ...........((((((((---------((.(((((....))(((((.-----...)))))........))))))))))))).....((((.((((......))))...))))....... ( -30.20) >DroSim_CAF1 125198 106 + 1 AUUCCCAUUCCCAUCCGCU---------UGUGCUGCUCUUGCAGAGCU-----UUUGUUCUACCCUUCUGGCCAAGUGGAUGUUAUAACUUGGUAAUUCGAUUUGCACAAAGUAUUUUCC ...........((((((((---------((.(((((....))(((((.-----...)))))........))))))))))))).....((((.((((......))))...))))....... ( -30.20) >DroEre_CAF1 138616 90 + 1 ----------------GCU---------CGUGCUGCUUUGGCAGUGCU-----UUUGUUCUACCCUUCUGGCCAAGUGGAUGUUAUAACUUGGUAAUUCGAUUUGCACAAAGUAUUUUCC ----------------...---------.(..((((....))))..)(-----(((((.(..........(((((((..........)))))))..........).))))))........ ( -21.85) >DroAna_CAF1 140962 118 + 1 --UACCGCUUCCUUCUGCUCUUCUUUUUUCUCCUUCUGCAGCAGGGCUUUGGCUUUGUUCUACCCUUCUGGCCAAGUGGAUGUUAUAACUUGGUAAUUUGAUUUGCACAAAGUAUUUUCC --....((..(((.((((...................)))).)))))....(((((((.(..........(((((((..........)))))))..........).)))))))....... ( -24.86) >consensus AUUCCCAUUCCCAUCCGCU_________UGUGCUGCUCUUGCAGAGCU_____UUUGUUCUACCCUUCUGGCCAAGUGGAUGUUAUAACUUGGUAAUUCGAUUUGCACAAAGUAUUUUCC ...........((((((((..........(..((((....))))..)................((....))...)))))))).....((((.((((......))))...))))....... (-15.78 = -17.26 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:23 2006