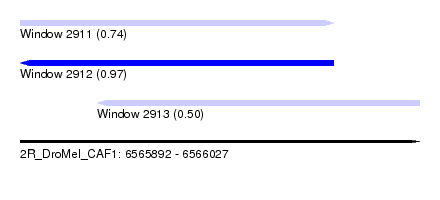
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,565,892 – 6,566,027 |
| Length | 135 |
| Max. P | 0.974228 |

| Location | 6,565,892 – 6,565,998 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -19.35 |
| Energy contribution | -17.88 |
| Covariance contribution | -1.47 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.44 |
| SVM RNA-class probability | 0.737350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
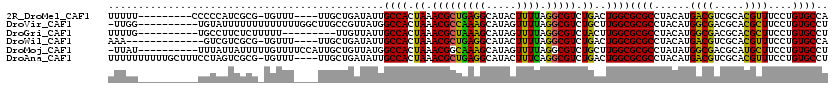
>2R_DroMel_CAF1 6565892 106 + 20766785 UUUUU---------CCCCCAUCGCG-UGUUU----UUGCUGAUAUUGCCACUAAACGCUGAGGCAUACUUUUAGGCGUCUGACUGGCGCGCCUACAUGACGUCGCACGUUUCCUGUGCCA .....---------......(((.(-(((((----..((.......))....)))))))))((((((....(((((((((....)).)))))))...((((.....))))...)))))). ( -27.80) >DroVir_CAF1 93375 109 + 1 -UUGG----------UGUAUUUUUUUUUUUUUGGCUUGCCGUUAUGGCCACUAAACGCCAAAGCAUAGUUUUAGGCGUCUGCUUGGCGCGCCUACAUGGCGACGCACGCUUCCUGUGCCU -((((----------(((.........((..(((((.........)))))..))))))))).((((((....((((((.......(((((((.....)))).))))))))).)))))).. ( -35.01) >DroGri_CAF1 86843 101 + 1 UUUUG----------UGCCUUCUCUUUUU---------UUGUUAUUGCCACUAAACGCUAAAGCAUAGUUUUAGGCGUCUACUUGGCGCGCCUACAUGGCGACGCACGCUUCCUGUGCCU ....(----------(((...........---------........((((.((.((((((((((...)))))).)))).))..))))(((((.....)))).)))))((.......)).. ( -26.80) >DroWil_CAF1 148303 102 + 1 AAA-------------GUCGUCGCG-UGUUU----UUGCUGAUAUUGCCACUAAACGCUGAGGCAUACUUUUAGGCGUCUGACUGGCGCGCCUACAUGACGUCGCACGUUUCCUGUGCCA ...-------------(.(((((((-(....----..((.......))......))))((((((...(.....)(((((.....))))))))).)).)))).)(((((.....))))).. ( -31.20) >DroMoj_CAF1 99236 109 + 1 -UUAU----------UUUAUUAUUUUUGUUUUCCAUUGCUGUUAUGGCCACUAAACGGCAAAGCAUAGUUUUAGGCGUCUGCUUGGCGCGCCUAUAUGGCGACGCAUGCUUCCUGUGCCU -....----------....................((((((((..(....)..)))))))).((((((...(((((....)))))(((((((.....)))).))).......)))))).. ( -31.20) >DroAna_CAF1 119187 115 + 1 UUUUUUUUUUGCUUUCCUAGUCGCG-UGUUU----UUGCUGAUAUUGCCACUAAACGCUGAGGCAUACUUUCAGGCGUCUGACUGGCGCGCCUACAUGACGUCGCACGUUUCCUGUGCCU ...................(.((((-(((..----...((((...((((.(........).)))).....))))(((((.....)))))....))))).)).)(((((.....))))).. ( -29.30) >consensus UUUUU__________UGUAUUCGCG_UGUUU____UUGCUGAUAUUGCCACUAAACGCUAAAGCAUACUUUUAGGCGUCUGACUGGCGCGCCUACAUGACGACGCACGCUUCCUGUGCCU ..............................................((((.((.(((((((((.....)))).))))).))..))))((((......((((.....))))....)))).. (-19.35 = -17.88 + -1.47)
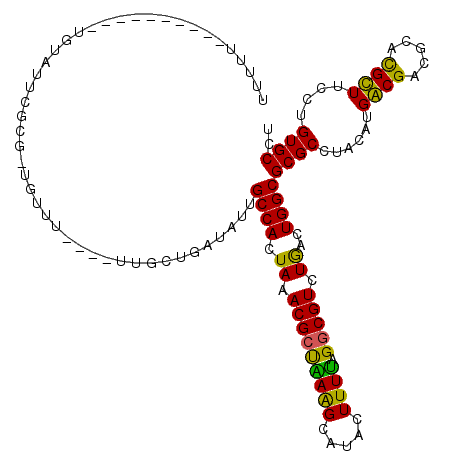

| Location | 6,565,892 – 6,565,998 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 76.13 |
| Mean single sequence MFE | -30.42 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.73 |
| SVM RNA-class probability | 0.974228 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6565892 106 - 20766785 UGGCACAGGAAACGUGCGACGUCAUGUAGGCGCGCCAGUCAGACGCCUAAAAGUAUGCCUCAGCGUUUAGUGGCAAUAUCAGCAA----AAACA-CGCGAUGGGGG---------AAAAA ..((((.(....)))))..((((...(((((((........).))))))...).)))(((((((((......((.......))..----....)-)))..))))).---------..... ( -29.10) >DroVir_CAF1 93375 109 - 1 AGGCACAGGAAGCGUGCGUCGCCAUGUAGGCGCGCCAAGCAGACGCCUAAAACUAUGCUUUGGCGUUUAGUGGCCAUAACGGCAAGCCAAAAAAAAAAAAAUACA----------CCAA- .(((((.....((..(((.((((.....)))))))...))(((((((..............))))))).)).(((.....)))..))).................----------....- ( -33.54) >DroGri_CAF1 86843 101 - 1 AGGCACAGGAAGCGUGCGUCGCCAUGUAGGCGCGCCAAGUAGACGCCUAAAACUAUGCUUUAGCGUUUAGUGGCAAUAACAA---------AAAAAGAGAAGGCA----------CAAAA ..((.......))(((((.((((.....)))))((((..(((((((.((((.......))))))))))).))))........---------...........)))----------).... ( -29.80) >DroWil_CAF1 148303 102 - 1 UGGCACAGGAAACGUGCGACGUCAUGUAGGCGCGCCAGUCAGACGCCUAAAAGUAUGCCUCAGCGUUUAGUGGCAAUAUCAGCAA----AAACA-CGCGACGAC-------------UUU ..((((.(....)))))..((((...(((((((........).)))))).............((((......((.......))..----....)-)))))))..-------------... ( -29.60) >DroMoj_CAF1 99236 109 - 1 AGGCACAGGAAGCAUGCGUCGCCAUAUAGGCGCGCCAAGCAGACGCCUAAAACUAUGCUUUGCCGUUUAGUGGCCAUAACAGCAAUGGAAAACAAAAAUAAUAAA----------AUAA- .((((....((((((((((((((.....)))((.....)).)))))........)))))))))).....((..((((.......))))...))............----------....- ( -29.20) >DroAna_CAF1 119187 115 - 1 AGGCACAGGAAACGUGCGACGUCAUGUAGGCGCGCCAGUCAGACGCCUGAAAGUAUGCCUCAGCGUUUAGUGGCAAUAUCAGCAA----AAACA-CGCGACUAGGAAAGCAAAAAAAAAA ..((((.(....)))))...(((.((.(((((..(...((((....))))..)..)))))))((((......((.......))..----....)-))))))................... ( -31.30) >consensus AGGCACAGGAAACGUGCGACGCCAUGUAGGCGCGCCAAGCAGACGCCUAAAACUAUGCCUCAGCGUUUAGUGGCAAUAACAGCAA____AAACA_AGAGAAGACA__________AAAAA ..((((.(....)))))...(((((.(((((((........).)))))).....((((....))))...))))).............................................. (-22.37 = -22.32 + -0.05)

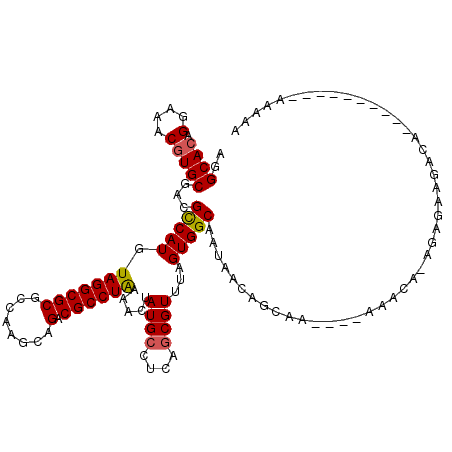
| Location | 6,565,918 – 6,566,027 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 72.70 |
| Mean single sequence MFE | -36.59 |
| Consensus MFE | -22.37 |
| Energy contribution | -22.32 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.17 |
| Structure conservation index | 0.61 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6565918 109 - 20766785 UCGCUUUUGUUUU--UUCUUGU---U------GCUCCUCCUGGCACAGGAAACGUGCGACGUCAUGUAGGCGCGCCAGUCAGACGCCUAAAAGUAUGCCUCAGCGUUUAGUGGCAAUAUC ((((...((((..--.((((((---.------(((......))))))))))))).)))).(((((.(((((((...((.((.((........)).)).))..))))))))))))...... ( -30.30) >DroVir_CAF1 93404 108 - 1 --GUUU-----UG--UUGCUUCGCCUUCGUAGUGG---CCAGGCACAGGAAGCGUGCGUCGCCAUGUAGGCGCGCCAAGCAGACGCCUAAAACUAUGCUUUGGCGUUUAGUGGCCAUAAC --....-----.(--(..((.((((..((((((((---(.(.((((.(....))))).).)))...(((((((........).))))))..))))))....))))...))..))...... ( -39.50) >DroGri_CAF1 86864 104 - 1 CGGCCC-----CG--GUCCCGGCCCA------GGC---CCAGGCACAGGAAGCGUGCGUCGCCAUGUAGGCGCGCCAAGUAGACGCCUAAAACUAUGCUUUAGCGUUUAGUGGCAAUAAC .((((.-----.(--((....)))..------)))---)...((((.(....)))))(.((((.....)))))((((..(((((((.((((.......))))))))))).))))...... ( -39.20) >DroWil_CAF1 148325 102 - 1 ------UUGGUGUUGCCCGCGU---U------GCU---GUUGGCACAGGAAACGUGCGACGUCAUGUAGGCGCGCCAGUCAGACGCCUAAAAGUAUGCCUCAGCGUUUAGUGGCAAUAUC ------..((((((((((....---(------(((---(..(((...(....)(((((....)...(((((((........).))))))...))))))).)))))....).))))))))) ( -37.00) >DroMoj_CAF1 99265 104 - 1 CGGUUU-----UU--GUGUUGUGGCU------GGC---CCAGGCACAGGAAGCAUGCGUCGCCAUAUAGGCGCGCCAAGCAGACGCCUAAAACUAUGCUUUGCCGUUUAGUGGCCAUAAC ......-----..--..(((((((((------...---...((((....((((((((((((((.....)))((.....)).)))))........)))))))))).......))))))))) ( -36.42) >DroAna_CAF1 119222 95 - 1 -----------GU--GGGGUGU---U------GUU---GCAGGCACAGGAAACGUGCGACGUCAUGUAGGCGCGCCAGUCAGACGCCUGAAAGUAUGCCUCAGCGUUUAGUGGCAAUAUC -----------..--..(((((---(------((.---.(..((((.(....)))))(((((..((.(((((..(...((((....))))..)..))))))))))))..)..)))))))) ( -37.10) >consensus __G_UU_____UU__GUCCUGU___U______GGU___CCAGGCACAGGAAACGUGCGACGCCAUGUAGGCGCGCCAAGCAGACGCCUAAAACUAUGCCUCAGCGUUUAGUGGCAAUAAC ..........................................((((.(....)))))...(((((.(((((((........).)))))).....((((....))))...)))))...... (-22.37 = -22.32 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:14 2006