
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
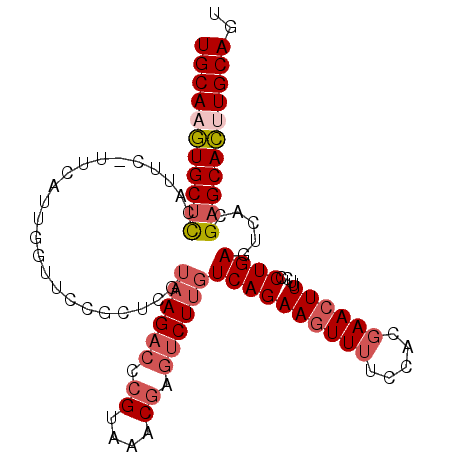
| Location | 6,562,057 – 6,562,160 |
| Length | 103 |
| Max. P | 0.955297 |

| Location | 6,562,057 – 6,562,160 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -30.32 |
| Consensus MFE | -25.20 |
| Energy contribution | -26.45 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.06 |
| Structure conservation index | 0.83 |
| SVM decision value | 1.45 |
| SVM RNA-class probability | 0.955297 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
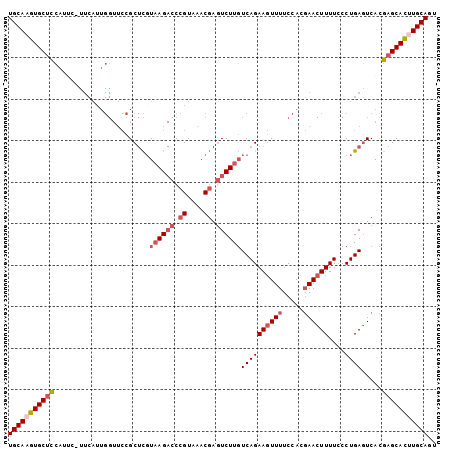
>2R_DroMel_CAF1 6562057 103 + 20766785 UGCAAGUGCUCCAUUCAUUCAUUGGUUCCGCUCGUAAGACCCGUAAACGAGUCUUGUCAGAAGUUUUCCACGAACUUUUGCCUGAGUCACGAGCACUUGCAGU (((((((((((.(((((..((..(((((..((..((((((.((....)).))))))..))..((.....)))))))..))..)))))...))))))))))).. ( -37.70) >DroSec_CAF1 114715 103 + 1 UGCAAGUGCUCCAUUCAUUCAUUGGUUCCGCUCCUAAGACCCGUAAACGAGUCUUGUCAGAAGUUUUCCACGAACUUUUCCCUGAGUCACGAGCACUUGCAGU (((((((((((.(((((......(((((..((..((((((.((....)).))))))..))..((.....)))))))......)))))...))))))))))).. ( -33.40) >DroSim_CAF1 100848 103 + 1 UGCAAGUGCUCCAUUCAUUCAUUGGUUCCGCUCGUAAGACCCGUAAACGAGUCUUGUCAGAAGUUUUCCACGAACUUUUCCCUGAGUCACGAGCACUUGCAGU (((((((((((.(((((......(((((..((..((((((.((....)).))))))..))..((.....)))))))......)))))...))))))))))).. ( -34.60) >DroEre_CAF1 115401 98 + 1 UGCA-GUGCUCCA----UUCAUUGGUUCCGCUCGUAAGACCCGUAAACGAGUCUUGUCAGAAGUUUUCCACGAACUUUUCCCUGAGUCACGAGCACUUGCAGC ((((-((((((((----.....)).....(((((((((((.((....)).)))))).)((((((((.....))))))))....))))...)))))).)))).. ( -29.50) >DroYak_CAF1 120498 99 + 1 UGCAAGUGCUUCA----UUCAUUGGUUCCGCUCGUAAGACCCGUAAACGAGUCUCGUCAGAAGUUUUCCACGAACUUUUCCCUGAGUCACGAGCACUUGCAGU (((((((((((.(----((((..((....((((((...........))))))......((((((((.....)))))))))).)))))...))))))))))).. ( -29.90) >DroAna_CAF1 115348 86 + 1 UGCAAUUGCUCGAUUC------------GGC-UGUAAG--CCGUAAACU-UUCUUCUCAGAACUUUUCCACUAA-UUUUCCCUGACUCAUGUGCAACUGCACU ((((.((((.((...(------------(((-.....)--)))......-......((((..............-......))))....)).)))).)))).. ( -16.85) >consensus UGCAAGUGCUCCAUUC_UUCAUUGGUUCCGCUCGUAAGACCCGUAAACGAGUCUUGUCAGAAGUUUUCCACGAACUUUUCCCUGAGUCACGAGCACUUGCAGU (((((((((((.......................((((((.((....)).))))))((((((((((.....))))))....)))).....))))))))))).. (-25.20 = -26.45 + 1.25)

| Location | 6,562,057 – 6,562,160 |
|---|---|
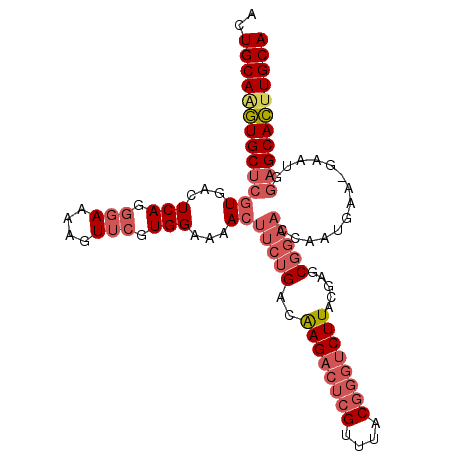
| Length | 103 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.83 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -27.88 |
| Energy contribution | -29.68 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.09 |
| Mean z-score | -3.48 |
| Structure conservation index | 0.79 |
| SVM decision value | 1.30 |
| SVM RNA-class probability | 0.939087 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
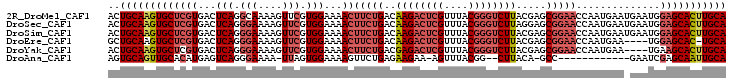
>2R_DroMel_CAF1 6562057 103 - 20766785 ACUGCAAGUGCUCGUGACUCAGGCAAAAGUUCGUGGAAAACUUCUGACAAGACUCGUUUACGGGUCUUACGAGCGGAACCAAUGAAUGAAUGGAGCACUUGCA ...((((((((((.....(((..((...(((((((((.....)))...((((((((....))))))))))))))(....)..))..)))...)))))))))). ( -38.60) >DroSec_CAF1 114715 103 - 1 ACUGCAAGUGCUCGUGACUCAGGGAAAAGUUCGUGGAAAACUUCUGACAAGACUCGUUUACGGGUCUUAGGAGCGGAACCAAUGAAUGAAUGGAGCACUUGCA ...((((((((((((...(((.(((....))).)))...))(((((.(((((((((....)))))))).)...)))))..............)))))))))). ( -37.60) >DroSim_CAF1 100848 103 - 1 ACUGCAAGUGCUCGUGACUCAGGGAAAAGUUCGUGGAAAACUUCUGACAAGACUCGUUUACGGGUCUUACGAGCGGAACCAAUGAAUGAAUGGAGCACUUGCA ...((((((((((.(.(.(((.((....(((((((((.....)))...((((((((....))))))))))))))....))..))).).)...)))))))))). ( -38.80) >DroEre_CAF1 115401 98 - 1 GCUGCAAGUGCUCGUGACUCAGGGAAAAGUUCGUGGAAAACUUCUGACAAGACUCGUUUACGGGUCUUACGAGCGGAACCAAUGAA----UGGAGCAC-UGCA ...(((.((((((.....(((.((....(((((((((.....)))...((((((((....))))))))))))))....))..))).----..))))))-))). ( -33.70) >DroYak_CAF1 120498 99 - 1 ACUGCAAGUGCUCGUGACUCAGGGAAAAGUUCGUGGAAAACUUCUGACGAGACUCGUUUACGGGUCUUACGAGCGGAACCAAUGAA----UGAAGCACUUGCA ...((((((((((((...(((.((....(((((((((.....)))...((((((((....))))))))))))))....))..))))----)).))))))))). ( -37.60) >DroAna_CAF1 115348 86 - 1 AGUGCAGUUGCACAUGAGUCAGGGAAAA-UUAGUGGAAAAGUUCUGAGAAGAA-AGUUUACGG--CUUACA-GCC------------GAAUCGAGCAAUUGCA ..(((((((((...(((.((........-...(((((....((((....))))-..)))))((--(.....-)))------------)).))).))))))))) ( -24.90) >consensus ACUGCAAGUGCUCGUGACUCAGGGAAAAGUUCGUGGAAAACUUCUGACAAGACUCGUUUACGGGUCUUACGAGCGGAACCAAUGAA_GAAUGGAGCACUUGCA ..(((((((((((((...(((.(((....))).)))...))(((((..((((((((....)))))))).....)))))..............))))))))))) (-27.88 = -29.68 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:12 2006