
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,550,739 – 6,550,853 |
| Length | 114 |
| Max. P | 0.997888 |

| Location | 6,550,739 – 6,550,832 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 91.40 |
| Mean single sequence MFE | -28.79 |
| Consensus MFE | -25.75 |
| Energy contribution | -25.95 |
| Covariance contribution | 0.20 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.89 |
| SVM decision value | 2.95 |
| SVM RNA-class probability | 0.997888 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
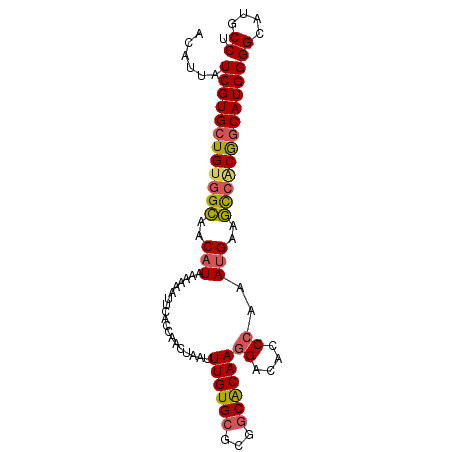
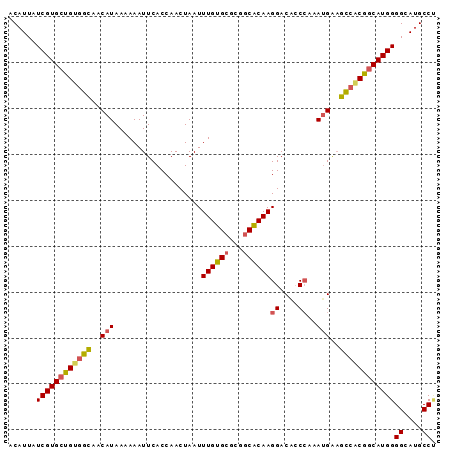
>2R_DroMel_CAF1 6550739 93 + 20766785 ACAUUAUCGUGCUGUGGCAACAUAAAAAAUUCACCAACUAAUUUGUGCGCCGCACAAGGACACCCAAAUGAAGCCACGGCAUGGGGCAUGCCU .......(((((((((((..(((...................((((((...))))))((....))..)))..)))))))))))(((....))) ( -31.50) >DroSec_CAF1 103480 93 + 1 ACAUUAUCGUGCUGUGGCAACAUAAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACAACCAAAUGAAGCCACGGCAUGGGGCAUGCCU .......(((((((((((..(((...................((((((...))))))((....))..)))..)))))))))))(((....))) ( -31.10) >DroSim_CAF1 89740 93 + 1 ACAUUAUCGUGCUGUGGCAACAUAAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACACCCAAAUGAAGCCACGGCAUGGGGCAUGCCC .......(((((((((((..(((...................((((((...))))))((....))..)))..)))))))))))(((....))) ( -32.80) >DroEre_CAF1 104352 93 + 1 ACAUUAUCGUGCUGUGGCAACAUAAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACGCCCAAAUGGAGCCACGGCAUGGGGCAUGCCU .......(((((((((((..(((...................((((((...))))))((....))..)))..)))))))))))(((....))) ( -32.30) >DroYak_CAF1 108846 93 + 1 ACAUUAUCGUGCUGUGGCAACAUAAAAAAUUCACCAACUAAUUUGUGCGCUUCACAAGGACACCCAAAUGAAGCAGCGGCAUGGGGCAUGCCU .......((((((((.(((.((.....................)))))((((((...((....))...)))))).))))))))(((....))) ( -24.60) >DroAna_CAF1 104029 93 + 1 ACAUUAUCGUGCUGUGGUAACAUAAAAAAUUCACCAACUAAUUUGUGCGAGGCGCAAGGACAACAGCAAGACAUCCCAACAUGGGGCAGGCCA ......((.(((((((((..............)))......(((((((...)))))))....)))))).))..((((.....))))....... ( -20.44) >consensus ACAUUAUCGUGCUGUGGCAACAUAAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACACCCAAAUGAAGCCACGGCAUGGGGCAUGCCU ......((((((((((((..(((...................((((((...))))))((....))..)))..))))))))))))((....)). (-25.75 = -25.95 + 0.20)

| Location | 6,550,762 – 6,550,853 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 71.30 |
| Mean single sequence MFE | -21.42 |
| Consensus MFE | -11.91 |
| Energy contribution | -12.05 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.38 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
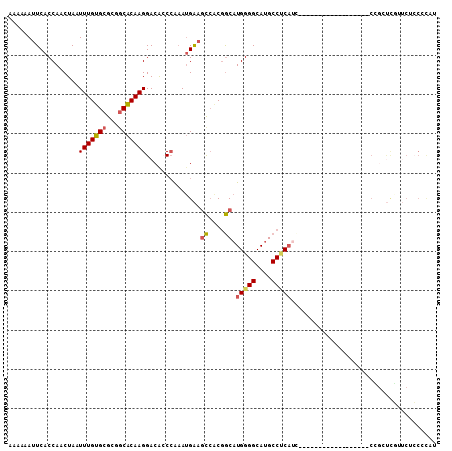
>2R_DroMel_CAF1 6550762 91 + 20766785 AAAAAAUUCACCAACUAAUUUGUGCGCCGCACAAGGACACCCAAAUGAAGCCACGGCAUGGGGCAUGCCUCAUU------------------CCGCUCGCUCACCGCAU ......((((.........((((((...))))))((....))...))))((..(((.((((((....)))))).------------------)))...))......... ( -22.30) >DroSec_CAF1 103503 91 + 1 AAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACAACCAAAUGAAGCCACGGCAUGGGGCAUGCCUCAUC------------------CCGCUCGUUCUCCCCAU ...................((((((...))))))(((.(((....((....))(((.((((((....)))))).------------------)))...))).))).... ( -20.80) >DroSim_CAF1 89763 109 + 1 AAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACACCCAAAUGAAGCCACGGCAUGGGGCAUGCCCCAUCCUACUCAACCUCCUCAUUCAGCUCGUUCUCCCCAU ...................((((((...))))))((((..(..(((((.((....))((((((....))))))..............)))))..)...))))....... ( -22.80) >DroEre_CAF1 104375 71 + 1 AAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACGCCCAAAUGGAGCCACGGCAUGGGGCAUGCCUC-------------------------------------- ..................(((((((...)))))))...((((....)).))...((((((...))))))..-------------------------------------- ( -17.50) >DroYak_CAF1 108869 89 + 1 AAAAAAUUCACCAACUAAUUUGUGCGCUUCACAAGGACACCCAAAUGAAGCAGCGGCAUGGGGCAUGCCUCAUC------------------CUCAUCA--CACCGCCU ....................((((.((((((...((....))...)))))).(.((.((((((....)))))))------------------).)..))--))...... ( -22.60) >DroAna_CAF1 104052 104 + 1 AAAAAAUUCACCAACUAAUUUGUGCGAGGCGCAAGGACAACAGCAAGACAUCCCAACAUGGGGCAGGCCACAAGACAC---ACCUCUCC--CCGGCCUCUUCUGCCUGC .......................((.(((((.(((...............((((.....)))).(((((...(((...---...)))..--..)))))))).))))))) ( -22.50) >consensus AAAAAAUUCACCAACUAAUUUGUGCGCGGCACAAGGACACCCAAAUGAAGCCACGGCAUGGGGCAUGCCUCAUC__________________CCGCUCGUUCUCCCCAU ..................(((((((...)))))))..............((....)).(((((....)))))..................................... (-11.91 = -12.05 + 0.14)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:12:05 2006