
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,511,791 – 6,511,888 |
| Length | 97 |
| Max. P | 0.818846 |

| Location | 6,511,791 – 6,511,888 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -35.32 |
| Consensus MFE | -20.40 |
| Energy contribution | -21.52 |
| Covariance contribution | 1.11 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818846 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
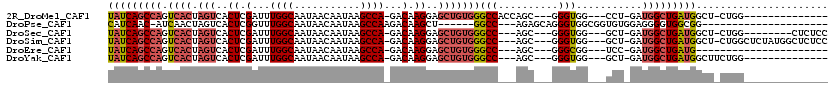
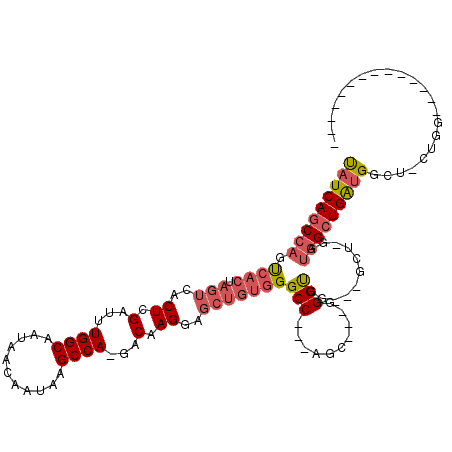
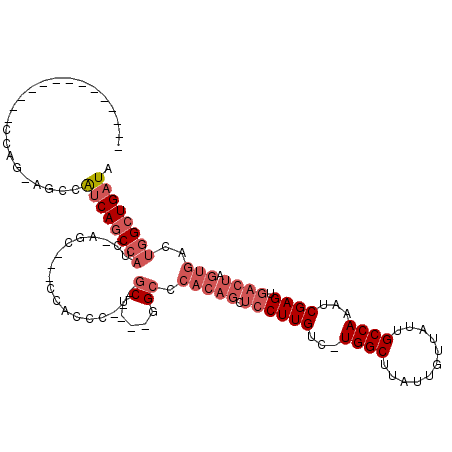
>2R_DroMel_CAF1 6511791 97 + 20766785 UAUCAGCCAGUCACUAGUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCCACCAGC---GGGUGG---CCU-GAUGGCUGAUGGCU-CUGG-------------- (((((((((..(((.(((..((.(.((((((...........))))-))).))..))))))(((((((...---.)))))---)).-..)))))))))...-....-------------- ( -42.30) >DroPse_CAF1 76210 89 + 1 CAUCAAC-AUCAACUAGUCACUCGGUUUGGCAAUAACAAUAAGCCAAGACAAGCU------GGCC---AGAGCAGGGUGGCGGUGUGGAGGGGUGGCGG--------------------- ((((...-.((.(((.((((((((.((((((..........(((........)))------.)))---))).).))))))))))...))..))))....--------------------- ( -26.90) >DroSec_CAF1 78931 100 + 1 UAUCAGCCAGUCACUAGUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCC---AGC---GGGUGG---GCU-GAUGGCUGAUGGCU-CUGG--------CUCUCC ....((((.(((...((((....))))((((...........))))-)))..((((((((.((((---(.(---((....---.))-).))))).))))))-))))--------)).... ( -37.10) >DroSim_CAF1 65311 108 + 1 UAUCAGCCAGUCACUAGUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCC---AGC---GGGUGG---GCU-GAUGGCUGAUGGCU-CUGGCUCUAUGGCUCUCC ....(((((......((((....(.((((((...........))))-)))..((((((((.((((---(.(---((....---.))-).))))).))))))-))))))...))))).... ( -39.00) >DroEre_CAF1 80576 87 + 1 UAUCAGCCAGUCACUAGUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCC---AGC---GGGCGG---UCC-GAUGGCUGAUG---------------------- (((((((((.((..((((..((.(.((((((...........))))-))).))..)))).(((((---...---....))---)))-)))))))))))---------------------- ( -33.10) >DroYak_CAF1 83139 95 + 1 UAUCAGCCAGUCACUAGUCACUCGAUUUGGCAAUAACAAUAAGCCA-GACAAGGAGCUGUGGGCC---AGC---GGGUGG---GCU-GAUGGCUGAUGGCUUCUGG-------------- (((((((((.((((.(((..((.(.((((((...........))))-))).))..)))))))..(---(((---......---)))-).)))))))))........-------------- ( -33.50) >consensus UAUCAGCCAGUCACUAGUCACUCGAUUUGGCAAUAACAAUAAGCCA_GACAAGGAGCUGUGGGCC___AGC___GGGUGG___GCU_GAUGGCUGAUGGCU_CUGG______________ (((((((((.((((.(((..((.(...((((...........))))...).))..)))))))(((..........)))...........)))))))))...................... (-20.40 = -21.52 + 1.11)
| Location | 6,511,791 – 6,511,888 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.01 |
| Mean single sequence MFE | -30.25 |
| Consensus MFE | -18.66 |
| Energy contribution | -20.52 |
| Covariance contribution | 1.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.633150 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
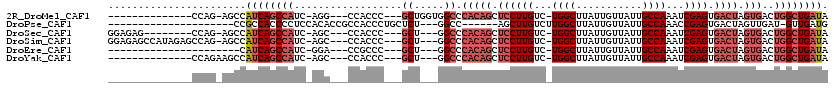
>2R_DroMel_CAF1 6511791 97 - 20766785 --------------CCAG-AGCCAUCAGCCAUC-AGG---CCACCC---GCUGGUGGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUA --------------....-....((((((((..-.((---(((((.---...)))))))(((((.((((((..-((((...........))))...)))).)))).)))..)))))))). ( -39.50) >DroPse_CAF1 76210 89 - 1 ---------------------CCGCCACCCCUCCACACCGCCACCCUGCUCU---GGCC------AGCUUGUCUUGGCUUAUUGUUAUUGCCAAACCGAGUGACUAGUUGAU-GUUGAUG ---------------------..........((.(((.(((((........)---))).------.(((((..(((((...........)))))..)))))........).)-)).)).. ( -18.50) >DroSec_CAF1 78931 100 - 1 GGAGAG--------CCAG-AGCCAUCAGCCAUC-AGC---CCACCC---GCU---GGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUA ((....--------))..-....((((((((((-(((---......---)))---))..(((((.((((((..-((((...........))))...)))).)))).)))..)))))))). ( -31.30) >DroSim_CAF1 65311 108 - 1 GGAGAGCCAUAGAGCCAG-AGCCAUCAGCCAUC-AGC---CCACCC---GCU---GGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUA ((...((......))...-..))((((((((((-(((---......---)))---))..(((((.((((((..-((((...........))))...)))).)))).)))..)))))))). ( -32.40) >DroEre_CAF1 80576 87 - 1 ----------------------CAUCAGCCAUC-GGA---CCGCCC---GCU---GGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUA ----------------------.((((((((((-((.---....))---(((---(.....))))((((((..-((((...........))))...)))).)).....)).)))))))). ( -29.20) >DroYak_CAF1 83139 95 - 1 --------------CCAGAAGCCAUCAGCCAUC-AGC---CCACCC---GCU---GGCCCACAGCUCCUUGUC-UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUA --------------.........((((((((((-(((---......---)))---))..(((((.((((((..-((((...........))))...)))).)))).)))..)))))))). ( -30.60) >consensus ______________CCAG_AGCCAUCAGCCAUC_AGC___CCACCC___GCU___GGCCCACAGCUCCUUGUC_UGGCUUAUUGUUAUUGCCAAAUCGAGUGACUAGUGACUGGCUGAUA .......................((((((((..................((.....)).(((((.((((((...((((...........))))...)))).)))).)))..)))))))). (-18.66 = -20.52 + 1.86)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:56 2006