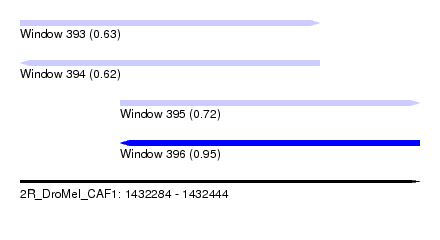
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,432,284 – 1,432,444 |
| Length | 160 |
| Max. P | 0.950141 |

| Location | 1,432,284 – 1,432,404 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -37.70 |
| Consensus MFE | -20.24 |
| Energy contribution | -27.18 |
| Covariance contribution | 6.94 |
| Combinations/Pair | 1.07 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627977 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1432284 120 + 20766785 ACGGCGAGGGACCAUCGAUAAUUCAUCCGGCUCCGCUAGCUGAGAGCCAUCCCGAUAAGAAUGUCCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAAUAAGGAGAUGGCCUCCAGCU ..((((.(((.((...(((.....))).)))))))))(((((((.((((((((((((....))))((((((((..((.((...)).))..)))))))).....)).)))))))).))))) ( -42.80) >DroSec_CAF1 50726 120 + 1 ACGGCGAGGGACCAUCGAUAAUCCAUCCGGCUCCGCUAGCUGAGAGCCAUCCCGAUAAGAAUGUCCAAGACAUAGUGAACAGUGUGNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNN .......((((.....(((.....))).(((((((.....)).))))).))))((((....))))....((((.(....).))))................................... ( -18.20) >DroEre_CAF1 48833 120 + 1 ACGGCGAAGGACCGUCGAUAAUCCAUCCGGCUCCGCUGGCUGAGAGUCAUCCCGACAAAAAUGUCCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAACAAGGAGAUGGCCUCUAGCU .(((((..(..(((..(((.....))))))..))))))((((((.((((((((((((....))))((((((((..((.((...)).))..)))))))).....)).))))))))).))). ( -42.40) >DroYak_CAF1 53899 120 + 1 ACGGCGAGGGACCGUCUAUUAUCCAUCCGGCUCCGCUGGCUGAGAGCCAUCCCGACAAGAAUGUUCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAAUAAGGAGAUGGCCUCCAGCU .(((((.(((.(((.............)))))))))))((((((.((((((((........((((((((((((..((.((...)).))..)))))))))))).)).)))))))).)))). ( -47.42) >consensus ACGGCGAGGGACCAUCGAUAAUCCAUCCGGCUCCGCUAGCUGAGAGCCAUCCCGACAAGAAUGUCCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAAUAAGGAGAUGGCCUCCAGCU .(((((.(((.(((..(((.....))))))))))))))((((((.((((((((((((....))))((((((((..((.((...)).))..)))))))).....)).))))))))).))). (-20.24 = -27.18 + 6.94)


| Location | 1,432,284 – 1,432,404 |
|---|---|
| Length | 120 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.69 |
| Mean single sequence MFE | -38.60 |
| Consensus MFE | -21.69 |
| Energy contribution | -28.25 |
| Covariance contribution | 6.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.09 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.621970 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1432284 120 - 20766785 AGCUGGAGGCCAUCUCCUUAUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGGACAUUCUUAUCGGGAUGGCUCUCAGCUAGCGGAGCCGGAUGAAUUAUCGAUGGUCCCUCGCCGU (((((..(((((((((..((((((((((((..((.((...)).))..))))))))))......))..)))))))))..))))).((((....((((.(........).))))....)))) ( -44.40) >DroSec_CAF1 50726 120 - 1 NNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNNCACACUGUUCACUAUGUCUUGGACAUUCUUAUCGGGAUGGCUCUCAGCUAGCGGAGCCGGAUGGAUUAUCGAUGGUCCCUCGCCGU .......................................((((((........)))))).......(((..(((((((........)))))))((.((((((....)))))).)).))). ( -23.40) >DroEre_CAF1 48833 120 - 1 AGCUAGAGGCCAUCUCCUUGUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGGACAUUUUUGUCGGGAUGACUCUCAGCCAGCGGAGCCGGAUGGAUUAUCGACGGUCCUUCGCCGU .(((((((..((((((......((((((((..((.((...)).))..))))))))((((....)))))))))).)))).)))..((((...((((.(((((......))))))))))))) ( -40.60) >DroYak_CAF1 53899 120 - 1 AGCUGGAGGCCAUCUCCUUAUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGAACAUUCUUGUCGGGAUGGCUCUCAGCCAGCGGAGCCGGAUGGAUAAUAGACGGUCCCUCGCCGU .((((..(((((((((.....(((((((((..((.((...)).))..)))))))))(((....))).)))))))))..))))..((((....((((.(........).))))....)))) ( -46.00) >consensus AGCUGGAGGCCAUCUCCUUAUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGGACAUUCUUAUCGGGAUGGCUCUCAGCCAGCGGAGCCGGAUGGAUUAUCGACGGUCCCUCGCCGU .(((((..((((((((......((((((((..((.((...)).))..))))))))((((....))))))))))))..)))))..((((....((((.(........).))))....)))) (-21.69 = -28.25 + 6.56)

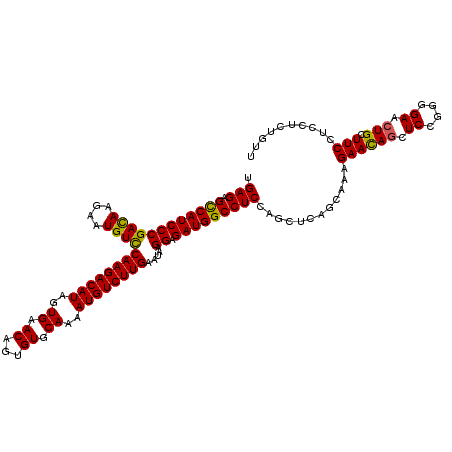
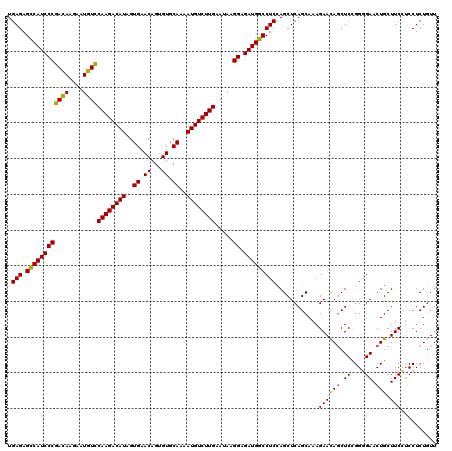
| Location | 1,432,324 – 1,432,444 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -35.47 |
| Consensus MFE | -31.38 |
| Energy contribution | -30.83 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718710 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1432324 120 + 20766785 UGAGAGCCAUCCCGAUAAGAAUGUCCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAAUAAGGAGAUGGCCUCCAGCUCAGCAAAGAAUAGCUCUGGGGAACUGCUUCUUCCUCUGUU .(((.((((((((((((....))))((((((((..((.((...)).))..)))))))).....)).))))))((((((...(((........)))))))))............))).... ( -37.00) >DroEre_CAF1 48873 120 + 1 UGAGAGUCAUCCCGACAAAAAUGUCCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAACAAGGAGAUGGCCUCUAGCUCAGCAAAGAACAGCUCCGGGGAACUGCUUCCACCUCUGUU .(((.((((((((((((....))))((((((((..((.((...)).))..)))))))).....)).))))))))).............(((((....((((((....)))).)).))))) ( -35.60) >DroYak_CAF1 53939 120 + 1 UGAGAGCCAUCCCGACAAGAAUGUUCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAAUAAGGAGAUGGCCUCCAGCUCAGCAAAGAACAACUCCGGCGAACUGCUUCCUCCUCUUUU .(((.((((((((........((((((((((((..((.((...)).))..)))))))))))).)).)))))))))(((.((.((...((.....))..)))).))).............. ( -33.80) >consensus UGAGAGCCAUCCCGACAAGAAUGUCCAAGACAUAGUGAACAGUGUGCAAAAUGUCUUGAAUAAGGAGAUGGCCUCCAGCUCAGCAAAGAACAGCUCCGGGGAACUGCUUCCUCCUCUGUU .(((.((((((((((((....))))((((((((..((.((...)).))..)))))))).....)).)))))))))............((((((.((....)).))).))).......... (-31.38 = -30.83 + -0.55)

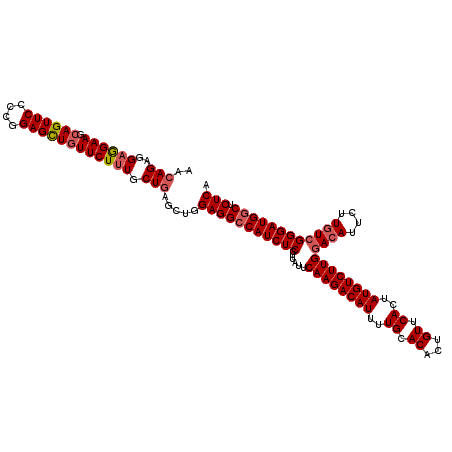
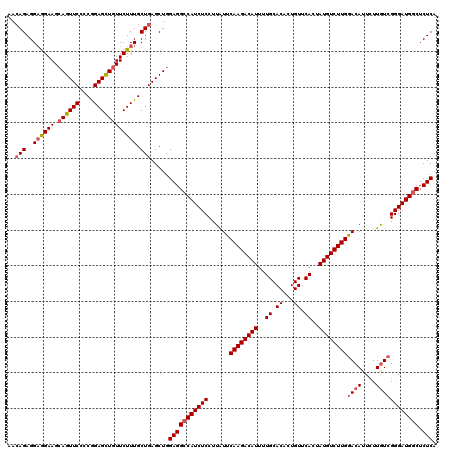
| Location | 1,432,324 – 1,432,444 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.78 |
| Mean single sequence MFE | -41.47 |
| Consensus MFE | -36.14 |
| Energy contribution | -37.70 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.05 |
| Structure conservation index | 0.87 |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.950141 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1432324 120 - 20766785 AACAGAGGAAGAAGCAGUUCCCCAGAGCUAUUCUUUGCUGAGCUGGAGGCCAUCUCCUUAUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGGACAUUCUUAUCGGGAUGGCUCUCA ..(((..((((((..(((((....))))).)))))).))).....(((((((((((..((((((((((((..((.((...)).))..))))))))))......))..)))))))).))). ( -40.70) >DroEre_CAF1 48873 120 - 1 AACAGAGGUGGAAGCAGUUCCCCGGAGCUGUUCUUUGCUGAGCUAGAGGCCAUCUCCUUGUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGGACAUUUUUGUCGGGAUGACUCUCA ...((.((..((((((((((....)))))))).))..))...))((((..((((((......((((((((..((.((...)).))..))))))))((((....)))))))))).)))).. ( -41.50) >DroYak_CAF1 53939 120 - 1 AAAAGAGGAGGAAGCAGUUCGCCGGAGUUGUUCUUUGCUGAGCUGGAGGCCAUCUCCUUAUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGAACAUUCUUGUCGGGAUGGCUCUCA ..............(((((((.(((((.....))))).)))))))(((((((((((.....(((((((((..((.((...)).))..)))))))))(((....))).)))))))).))). ( -42.20) >consensus AACAGAGGAGGAAGCAGUUCCCCGGAGCUGUUCUUUGCUGAGCUGGAGGCCAUCUCCUUAUUCAAGACAUUUUGCACACUGUUCACUAUGUCUUGGACAUUCUUGUCGGGAUGGCUCUCA ..(((..((((((.((((((....)))))))))))).))).....(((((((((((......((((((((..((.((...)).))..))))))))((((....)))))))))))).))). (-36.14 = -37.70 + 1.56)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:31:12 2006