
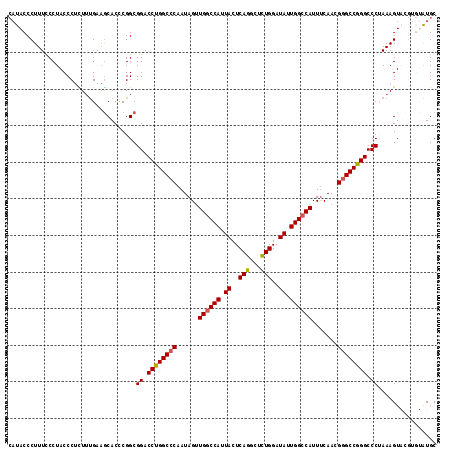
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,501,934 – 6,502,051 |
| Length | 117 |
| Max. P | 0.995642 |

| Location | 6,501,934 – 6,502,051 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -37.80 |
| Consensus MFE | -32.20 |
| Energy contribution | -32.20 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.85 |
| SVM decision value | 2.60 |
| SVM RNA-class probability | 0.995642 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
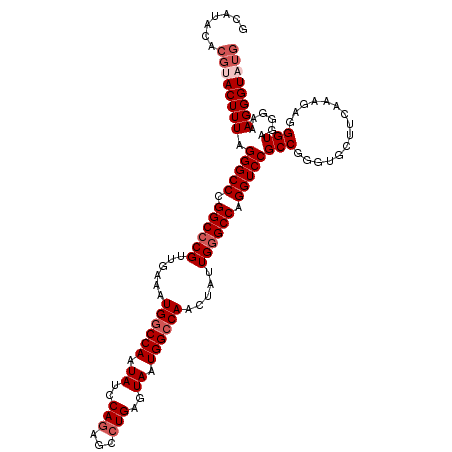
>2R_DroMel_CAF1 6501934 117 + 20766785 CAUACCCUUUUCCUACCCUCUUUGAAGCACCCGGCGGACCUGGCCCAAUAGUUGCCCAUUACUCAGGCUUUGGAUAUUGGCCAUUUCAACGGGCCGGGCCCUAAAGUACGUGUAUGC (((((.(...................((.....))((.((((((((...(((........)))..((((.........))))........)))))))).))........).))))). ( -30.90) >DroSec_CAF1 69212 117 + 1 CAUACCCUUUCCCUACCCUCUUUGAAGCACCCGGCGGACCUGGCCCAAUAGUUGGCCAUUACUCAGACUCUGGAUAUUGGCCAUUUCAACGGGCCGGGCCCUAAAGUACGUGUAUGC (((((.(...................((.....))((.((((((((......((((((.((..(((...)))..)).)))))).......)))))))).))........).))))). ( -39.52) >DroSim_CAF1 55555 117 + 1 CAUACCCUUUCCCUACCCUCUUUGAAGCACCCGGCGGACCUGGCCCAAUAGUUGGCCAUUACUCAGGCUCUGGAUAUUGGCCAUUUCAACGGGCCGGGCCCUAAAGUACGUGUAUGC (((((.(...................((.....))((.((((((((......((((((.((..(((...)))..)).)))))).......)))))))).))........).))))). ( -39.52) >DroEre_CAF1 71421 111 + 1 CAUACCCUUUCCCUACCCUUUGCGAAGCACCCGGCGGACCUGGCCCAAUAGUUGGCCAUUACUCAGGCUCUGGAUAUUGGCCAUUCCAACGGGCCAGGCCCAAAAGU------GUGC (((((..........((...(((...)))...)).((.((((((((......((((((.((..(((...)))..)).)))))).......)))))))).))....))------))). ( -40.92) >DroYak_CAF1 72801 108 + 1 --UACCCUUUCCCUACCCUUUUUGCCACACUCGGCGGACCUGGC-CAAUAGCUGGCCAUUACUCAGGCUCUGGAUAUUGGCCAUUUCAACGGGCCAGGCCCAAAAGU------AUGC --...............(((((.(((......)))((.((((((-(......((((((.((..(((...)))..)).))))))........))))))).))))))).------.... ( -38.14) >consensus CAUACCCUUUCCCUACCCUCUUUGAAGCACCCGGCGGACCUGGCCCAAUAGUUGGCCAUUACUCAGGCUCUGGAUAUUGGCCAUUUCAACGGGCCGGGCCCUAAAGUACGUGUAUGC ...................................((.((((((((......((((((.((..(((...)))..)).)))))).......)))))))).))................ (-32.20 = -32.20 + -0.00)


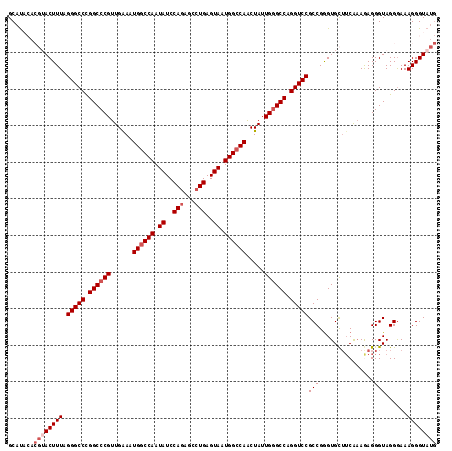
| Location | 6,501,934 – 6,502,051 |
|---|---|
| Length | 117 |
| Sequences | 5 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 89.86 |
| Mean single sequence MFE | -44.94 |
| Consensus MFE | -35.94 |
| Energy contribution | -37.44 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.80 |
| SVM decision value | 2.05 |
| SVM RNA-class probability | 0.986739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6501934 117 - 20766785 GCAUACACGUACUUUAGGGCCCGGCCCGUUGAAAUGGCCAAUAUCCAAAGCCUGAGUAAUGGGCAACUAUUGGGCCAGGUCCGCCGGGUGCUUCAAAGAGGGUAGGAAAAGGGUAUG .(((((.(.((((((((.(((((((.....((..(((((......((.....))..((((((....)))))))))))..)).))))))).))......)))))).)......))))) ( -38.50) >DroSec_CAF1 69212 117 - 1 GCAUACACGUACUUUAGGGCCCGGCCCGUUGAAAUGGCCAAUAUCCAGAGUCUGAGUAAUGGCCAACUAUUGGGCCAGGUCCGCCGGGUGCUUCAAAGAGGGUAGGGAAAGGGUAUG .(((((..((((((..(((((.((((((......((((((.((..(((...)))..)).)))))).....)))))).)))))...))))))(((............)))...))))) ( -44.30) >DroSim_CAF1 55555 117 - 1 GCAUACACGUACUUUAGGGCCCGGCCCGUUGAAAUGGCCAAUAUCCAGAGCCUGAGUAAUGGCCAACUAUUGGGCCAGGUCCGCCGGGUGCUUCAAAGAGGGUAGGGAAAGGGUAUG .(((((..((((((..(((((.((((((......((((((.((..(((...)))..)).)))))).....)))))).)))))...))))))(((............)))...))))) ( -44.30) >DroEre_CAF1 71421 111 - 1 GCAC------ACUUUUGGGCCUGGCCCGUUGGAAUGGCCAAUAUCCAGAGCCUGAGUAAUGGCCAACUAUUGGGCCAGGUCCGCCGGGUGCUUCGCAAAGGGUAGGGAAAGGGUAUG ((((------.((..(((((((((((((.(((..((((((.((..(((...)))..)).)))))).))).)))))))))))))..))))))(((.(........).)))........ ( -51.30) >DroYak_CAF1 72801 108 - 1 GCAU------ACUUUUGGGCCUGGCCCGUUGAAAUGGCCAAUAUCCAGAGCCUGAGUAAUGGCCAGCUAUUG-GCCAGGUCCGCCGAGUGUGGCAAAAAGGGUAGGGAAAGGGUA-- ...(------((((((((((((((((.((.(...((((((.((..(((...)))..)).)))))).).)).)-)))))))))((((....))))...)))))))...........-- ( -46.30) >consensus GCAUACACGUACUUUAGGGCCCGGCCCGUUGAAAUGGCCAAUAUCCAGAGCCUGAGUAAUGGCCAACUAUUGGGCCAGGUCCGCCGGGUGCUUCAAAGAGGGUAGGGAAAGGGUAUG .......((((((((.(((((.((((((......((((((.((..(((...)))..)).)))))).....)))))).)))))(((...............)))......)))))))) (-35.94 = -37.44 + 1.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:53 2006