| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,500,642 – 6,500,741 |
| Length | 99 |
| Max. P | 0.878588 |

| Location | 6,500,642 – 6,500,741 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -20.98 |
| Consensus MFE | -15.65 |
| Energy contribution | -15.52 |
| Covariance contribution | -0.14 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.75 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.878588 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
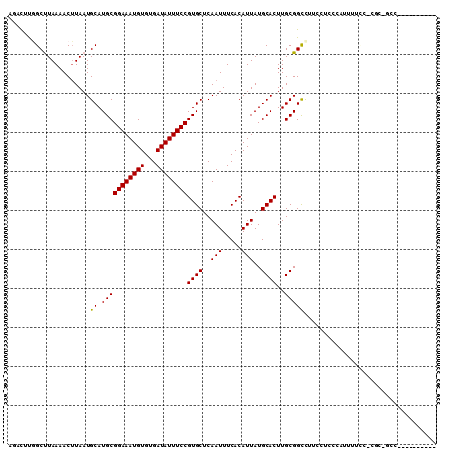
>2R_DroMel_CAF1 6500642 99 + 20766785 AGACUUGGCUUAAAACUUAAUGCAUGCGGAAAUGUGUGAUAUUUCCGUGCUCAAUUUCACAUUAUGCACUUGCGGCUUUCCUCCCAUUUUCC-CGCUGCC---------- ......(((...........((((((.((((((((...))))))))(((........)))..))))))...((((................)-))).)))---------- ( -20.89) >DroVir_CAF1 48960 78 + 1 AUAUACAGCCUAAAACUUAAUGCAUGCGGAAAUGUGUGAUAUUUCCGUGCUCAAUUUCACAUUAUGCACUUGCGGCCA-------------------------------- .......(((...........((..((((((((((...))))))))))))...............((....)))))..-------------------------------- ( -16.90) >DroEre_CAF1 70080 99 + 1 GGACUUGGCUUAAAACGUAAUGCAUGCGGAAAUGUGUGAUAUUUCCGUGCUCAAUUUCACAUUAUGCACUUGCGGCUUUCCUCCCAUUUUCC-CGCAGCC---------- (((..(((...((..(((((((((((.((((((((...))))))))(((........)))..)))))).)))))..)).....)))...)))-.......---------- ( -23.90) >DroWil_CAF1 87339 109 + 1 UGGCUUGGCUUAAAACUUAAUGCAUGCGGAAAUGUGUGAUAUUUCCGUGCUCAAUUUCACAUUAUGCACUUGCGGCCUCCGUUUGCCUUUUC-CUCUGCCACUUCUUCUC ((((..((.............(((...((((((((...))))))))((((..(((.....)))..)))).)))(((........)))....)-)...))))......... ( -24.70) >DroMoj_CAF1 55969 78 + 1 AUACACAGCUUAAAACUUAAUGCAUGCGGAAAUGUGUGAUAUUUCCGUGCUCAAUUUCACAUUAUGCACUUGCGGCCA-------------------------------- .....................((.(((((((((((...))))))))((((..(((.....)))..))))..)))))..-------------------------------- ( -15.70) >DroAna_CAF1 67831 100 + 1 AUACUUGGCUUAAAACUUAAUGCAUGCGGAAAUGUGUGAUAUUUCCGUGCUCAAUUUCACAUUAUGCACUUGCGGUUUUCCUCCCAUUUUCCGCGCCUUC---------- ......(((...........((((((.((((((((...))))))))(((........)))..))))))...((((...............)))))))...---------- ( -23.76) >consensus AGACUUGGCUUAAAACUUAAUGCAUGCGGAAAUGUGUGAUAUUUCCGUGCUCAAUUUCACAUUAUGCACUUGCGGCCUUCCUCCCAUUUUCC_CGC_GCC__________ .....................((.(((((((((((...))))))))((((..(((.....)))..))))..))))).................................. (-15.65 = -15.52 + -0.14)


| Location | 6,500,642 – 6,500,741 |
|---|---|
| Length | 99 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 77.29 |
| Mean single sequence MFE | -23.66 |
| Consensus MFE | -15.80 |
| Energy contribution | -15.80 |
| Covariance contribution | -0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.845316 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
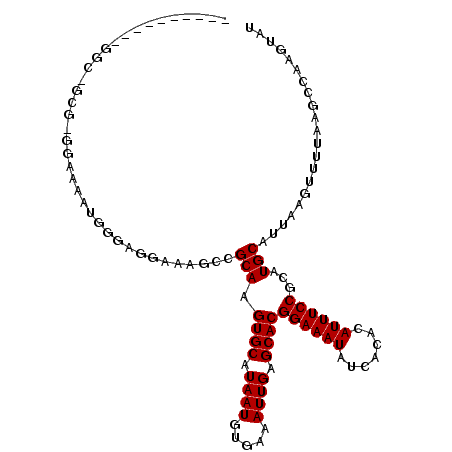
>2R_DroMel_CAF1 6500642 99 - 20766785 ----------GGCAGCG-GGAAAAUGGGAGGAAAGCCGCAAGUGCAUAAUGUGAAAUUGAGCACGGAAAUAUCACACAUUUCCGCAUGCAUUAAGUUUUAAGCCAAGUCU ----------(((.(((-(................)))).(((((((...(((........)))((((((.......))))))..))))))).........)))...... ( -24.89) >DroVir_CAF1 48960 78 - 1 --------------------------------UGGCCGCAAGUGCAUAAUGUGAAAUUGAGCACGGAAAUAUCACACAUUUCCGCAUGCAUUAAGUUUUAGGCUGUAUAU --------------------------------.((((((..((((.((((.....)))).))))((((((.......))))))))..((.....))....))))...... ( -19.80) >DroEre_CAF1 70080 99 - 1 ----------GGCUGCG-GGAAAAUGGGAGGAAAGCCGCAAGUGCAUAAUGUGAAAUUGAGCACGGAAAUAUCACACAUUUCCGCAUGCAUUACGUUUUAAGCCAAGUCC ----------((((...-..((((((...((....))(((.((((.((((.....)))).))))((((((.......))))))...)))....)))))).))))...... ( -26.70) >DroWil_CAF1 87339 109 - 1 GAGAAGAAGUGGCAGAG-GAAAAGGCAAACGGAGGCCGCAAGUGCAUAAUGUGAAAUUGAGCACGGAAAUAUCACACAUUUCCGCAUGCAUUAAGUUUUAAGCCAAGCCA .........((((..((-((...(((........)))(((.((((.((((.....)))).))))((((((.......))))))...)))......))))..))))..... ( -26.80) >DroMoj_CAF1 55969 78 - 1 --------------------------------UGGCCGCAAGUGCAUAAUGUGAAAUUGAGCACGGAAAUAUCACACAUUUCCGCAUGCAUUAAGUUUUAAGCUGUGUAU --------------------------------.....((..((((.((((.....)))).))))((((((.......))))))))((((((..(((.....))))))))) ( -18.80) >DroAna_CAF1 67831 100 - 1 ----------GAAGGCGCGGAAAAUGGGAGGAAAACCGCAAGUGCAUAAUGUGAAAUUGAGCACGGAAAUAUCACACAUUUCCGCAUGCAUUAAGUUUUAAGCCAAGUAU ----------...(((((((...............)))).(((((((...(((........)))((((((.......))))))..))))))).........)))...... ( -24.96) >consensus __________GGC_GCG_GGAAAAUGGGAGGAAAGCCGCAAGUGCAUAAUGUGAAAUUGAGCACGGAAAUAUCACACAUUUCCGCAUGCAUUAAGUUUUAAGCCAAGUAU .....................................(((.((((.((((.....)))).))))((((((.......))))))...)))..................... (-15.80 = -15.80 + -0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:50 2006