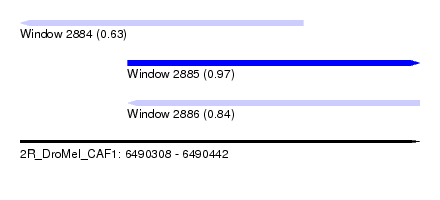
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,490,308 – 6,490,442 |
| Length | 134 |
| Max. P | 0.968088 |

| Location | 6,490,308 – 6,490,403 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 87.53 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -22.83 |
| Energy contribution | -23.47 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630472 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
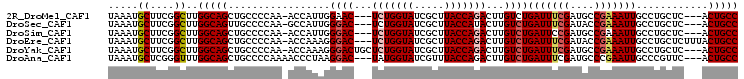
>2R_DroMel_CAF1 6490308 95 - 20766785 C---UCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUGCCGAAAUUGCCUGCUC---ACUGCCAAUGGCAUCAGGCAGUGCAC-UGCAAUAUGAAACUUA .---..((((((((.....((((....))))....))))))))..(((((..(..(---((((((..........)))))))..)-.))))).......... ( -30.00) >DroSec_CAF1 58096 95 - 1 C---UCUGGUAUCGCUUACCAUACUUGUCUGAUUUCGAUACCGAAAUUGCCUGCUC---ACUGCCAAUGGCAUCAGGCAGUGCAC-UGCAAGAGGAAACUUA .---..(((((.....)))))..(((((..(((((((....)))))))(((((((.---..((((...))))...))))).))..-.)))))((....)).. ( -30.20) >DroSim_CAF1 45618 95 - 1 C---UCUGGUAUCGCUUACCAGACUUGUCUGAUUCCGAUGCCGAAAUUGCCUGCUC---ACUGCCAAUGGCAUCAGGCAGUGCAC-UGCAAUAUGCAACUUA .---..((((((((.....((((....))))....))))))))....((((((((.---..((((...))))...))))).))).-(((.....)))..... ( -30.80) >DroEre_CAF1 56602 98 - 1 C---UCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUACCGAAAUUGCCUGCUCUUUACUGCCAAUGGCAUCAGGCAGUGCAC-UGCAAUGUGCAACUUA .---(((((((.....))))))).(((((((((((((....))))).((((.((........))....))))))))))))(((((-......)))))..... ( -34.40) >DroYak_CAF1 61480 98 - 1 CUGCUCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUGCCGAAAUUGCCUGCUC---ACUGCCAAUGGCAUCAGGCAGUGCAC-UGCAAUGUGAAACUUU .(((..((((((((.....((((....))))....))))))))....((((((((.---..((((...))))...))))).))).-.)))............ ( -31.20) >DroAna_CAF1 57520 94 - 1 C---UAUGGUAUCGUUUACCAGACUUGUCUGAUUUCGAUGCCCGAAUUGCCCGUUC---ACUGCCAGUGGCGGCUGUGACUACACGUGAAAAAAAAAAC--A .---...(((((((.....((((....))))....)))))))...........(((---((.(((......))).(((....)))))))).........--. ( -24.30) >consensus C___UCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUGCCGAAAUUGCCUGCUC___ACUGCCAAUGGCAUCAGGCAGUGCAC_UGCAAUAUGAAACUUA .......(((((((.....((((....))))....)))))))...((((((((........((((...)))).))))))))((....))............. (-22.83 = -23.47 + 0.64)

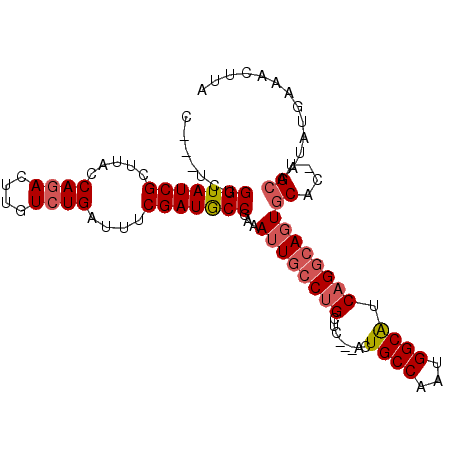
| Location | 6,490,344 – 6,490,442 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -35.07 |
| Consensus MFE | -30.01 |
| Energy contribution | -29.77 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.62 |
| SVM RNA-class probability | 0.968088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
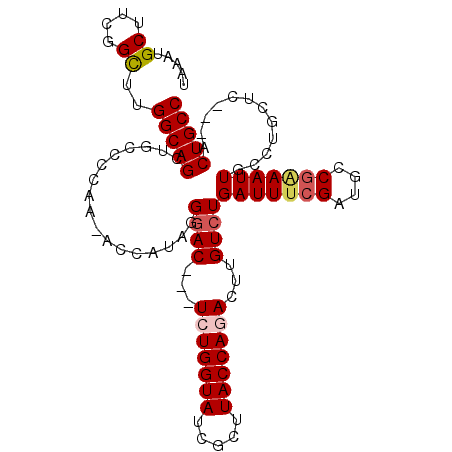
>2R_DroMel_CAF1 6490344 98 + 20766785 GGCAGU---GAGCAGGCAAUUUCGGCAUCGAAAUCAGACAAGUCUGGUAAGCGAUACCAGA---GUUCCAAUGGU-UUGGGGCAGCUGCCAAGCCGAAGCAUUUA (((..(---(.((((.(......((.((((..((((((....))))))...)))).))...---(((((((....-))))))).))))))).))).......... ( -33.90) >DroSec_CAF1 58132 98 + 1 GGCAGU---GAGCAGGCAAUUUCGGUAUCGAAAUCAGACAAGUAUGGUAAGCGAUACCAGA---GUCCCAAUGGC-UUGGGGCAACUGCCAAGCCGAAGCAUUUA (((..(---(.((((........(((((((..((((........))))...)))))))...---(((((((....-)))))))..)))))).))).......... ( -35.90) >DroSim_CAF1 45654 98 + 1 GGCAGU---GAGCAGGCAAUUUCGGCAUCGGAAUCAGACAAGUCUGGUAAGCGAUACCAGA---GUCCCAAUGGU-UUGGGGCAGCUGCCAAGCCGAAGCAUUUA (((..(---(.((((.(......((.((((..((((((....))))))...)))).))...---(((((((....-))))))).))))))).))).......... ( -35.60) >DroEre_CAF1 56638 101 + 1 GGCAGUAAAGAGCAGGCAAUUUCGGUAUCGAAAUCAGACAAGUCUGGUAAGCGAUACCAGA---GUCCCUUUGGU-UUGGGGCAGCUGCCAAGCCGAAGCAUUUA ((((((.................(((((((..((((((....))))))...)))))))...---(((((......-..))))).))))))..((....))..... ( -38.80) >DroYak_CAF1 61516 101 + 1 GGCAGU---GAGCAGGCAAUUUCGGCAUCGAAAUCAGACAAGUCUGGUAAGCGAUACCAGAGCAGUCCCUUUGGU-UUGGGGCAGCUGCCAAGCCGAAGCAUUUA .((..(---(.((.(((......((.((((..((((((....))))))...)))).))..(((.(((((......-..))))).))))))..))))..))..... ( -35.60) >DroAna_CAF1 57555 99 + 1 GGCAGU---GAACGGGCAAUUCGGGCAUCGAAAUCAGACAAGUCUGGUAAACGAUACCAUA---GUCCUUAGGGUUUUGGGGCAGCUGCCAAACCCGAGCAUUUA .((...---....((((......((.((((..((((((....))))))...)))).))...---))))...((((((...(((....)))))))))..))..... ( -30.60) >consensus GGCAGU___GAGCAGGCAAUUUCGGCAUCGAAAUCAGACAAGUCUGGUAAGCGAUACCAGA___GUCCCAAUGGU_UUGGGGCAGCUGCCAAGCCGAAGCAUUUA ((((((.................((.((((..((((((....))))))...)))).))......(((((.........))))).))))))............... (-30.01 = -29.77 + -0.25)


| Location | 6,490,344 – 6,490,442 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 90.70 |
| Mean single sequence MFE | -30.03 |
| Consensus MFE | -25.51 |
| Energy contribution | -26.10 |
| Covariance contribution | 0.59 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843296 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6490344 98 - 20766785 UAAAUGCUUCGGCUUGGCAGCUGCCCCAA-ACCAUUGGAAC---UCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUGCCGAAAUUGCCUGCUC---ACUGCC .....((....))..(((((..((.((((-....))))...---..((((((((.....((((....))))....)))))))).........))..---.))))) ( -29.20) >DroSec_CAF1 58132 98 - 1 UAAAUGCUUCGGCUUGGCAGUUGCCCCAA-GCCAUUGGGAC---UCUGGUAUCGCUUACCAUACUUGUCUGAUUUCGAUACCGAAAUUGCCUGCUC---ACUGCC .....((....))..((((((.(((((((-....)))))..---..(((((.....))))).........(((((((....)))))))....))..---)))))) ( -30.00) >DroSim_CAF1 45654 98 - 1 UAAAUGCUUCGGCUUGGCAGCUGCCCCAA-ACCAUUGGGAC---UCUGGUAUCGCUUACCAGACUUGUCUGAUUCCGAUGCCGAAAUUGCCUGCUC---ACUGCC .....((....))..(((((..(((((((-....)))))..---..((((((((.....((((....))))....)))))))).........))..---.))))) ( -32.50) >DroEre_CAF1 56638 101 - 1 UAAAUGCUUCGGCUUGGCAGCUGCCCCAA-ACCAAAGGGAC---UCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUACCGAAAUUGCCUGCUCUUUACUGCC .....((....))..(((((....(((..-......)))..---..((((((((.....((((....))))....)))))))).................))))) ( -29.00) >DroYak_CAF1 61516 101 - 1 UAAAUGCUUCGGCUUGGCAGCUGCCCCAA-ACCAAAGGGACUGCUCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUGCCGAAAUUGCCUGCUC---ACUGCC .....((....))..(((((..((.....-.......((((...(((((((.....)))))))...))))(((((((....)))))))....))..---.))))) ( -32.20) >DroAna_CAF1 57555 99 - 1 UAAAUGCUCGGGUUUGGCAGCUGCCCCAAAACCCUAAGGAC---UAUGGUAUCGUUUACCAGACUUGUCUGAUUUCGAUGCCCGAAUUGCCCGUUC---ACUGCC .....((..(((((((((....)))...))))))...((((---...(((((((.....((((....))))....))))))).(.......)))))---...)). ( -27.30) >consensus UAAAUGCUUCGGCUUGGCAGCUGCCCCAA_ACCAUAGGGAC___UCUGGUAUCGCUUACCAGACUUGUCUGAUUUCGAUGCCGAAAUUGCCUGCUC___ACUGCC .....((....))..(((((.................((((...(((((((.....)))))))...))))(((((((....)))))))............))))) (-25.51 = -26.10 + 0.59)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:48 2006