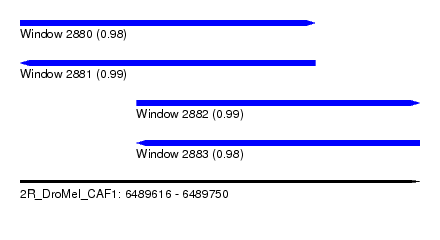
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,489,616 – 6,489,750 |
| Length | 134 |
| Max. P | 0.994695 |

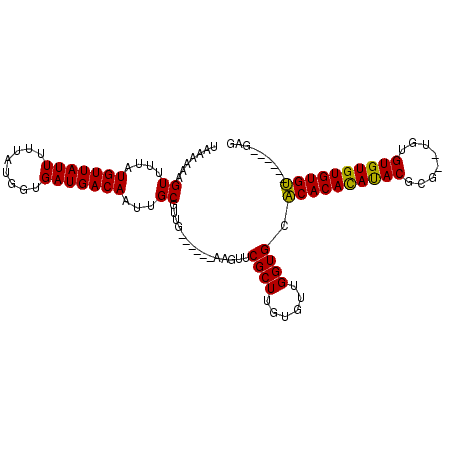
| Location | 6,489,616 – 6,489,715 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -27.15 |
| Consensus MFE | -21.70 |
| Energy contribution | -20.95 |
| Covariance contribution | -0.75 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.80 |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.984330 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6489616 99 + 20766785 UAAAAAAGUUUUAUGUUAUUUUUAUGGUGAUGACAAUUGCGUUG------AAGUUCGCUUGUGUUGGUGCACACACAUACGCG--UGUGUGUGUGUGUGUGAGAGAG ..............((((((.....))))))((((...(((...------.....)))...))))....(((((((((((((.--...)))))))))))))...... ( -28.30) >DroSec_CAF1 57350 92 + 1 UAAAAAAGUUUUAUGUUAUUUUUAUGGUGAUGACAAUUGCGUUG------AAGUUCGCUUGUGUUGGUGCACACAUAUACUA---CUCGUGUGUGUGU------GAG ..............((((((.....))))))((((...(((...------.....)))...))))....(((((((((((..---...))))))))))------).. ( -22.80) >DroSim_CAF1 44870 93 + 1 UAAAAAAGUUUUAUGUUAUUUUUAUGGUGAUGACAAUUGCGUUG------AAGUUCGCUUGUGUUGGUGCACACACAUACUCG--UUUGUGUGUGUGU------GAG ..............((((((.....))))))((((...(((...------.....)))...))))....(((((((((((...--...))))))))))------).. ( -25.50) >DroYak_CAF1 60682 101 + 1 UAAAAAAGUUUUAUGUUAUUUUUAUGGUGAUGACAAUUGCGUUUGCGUUCGCGUUCGCUUGUGUUGGUGGGCACACACACGUAUAUGUGUGUGUGUGC------GGG ..............((((((.....))))))((((...(((..(((....)))..)))...)))).....(((((((((((......)))))))))))------... ( -32.00) >consensus UAAAAAAGUUUUAUGUUAUUUUUAUGGUGAUGACAAUUGCGUUG______AAGUUCGCUUGUGUUGGUGCACACACAUACGCG__UGUGUGUGUGUGU______GAG .......((....(((((((........)))))))...))...............((((......)))).((((((((((........))))))))))......... (-21.70 = -20.95 + -0.75)

| Location | 6,489,616 – 6,489,715 |
|---|---|
| Length | 99 |
| Sequences | 4 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.50 |
| Mean single sequence MFE | -16.20 |
| Consensus MFE | -11.19 |
| Energy contribution | -11.07 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.08 |
| Mean z-score | -3.20 |
| Structure conservation index | 0.69 |
| SVM decision value | 2.07 |
| SVM RNA-class probability | 0.987085 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6489616 99 - 20766785 CUCUCUCACACACACACACACA--CGCGUAUGUGUGUGCACCAACACAAGCGAACUU------CAACGCAAUUGUCAUCACCAUAAAAAUAACAUAAAACUUUUUUA ........((((((((.((...--...)).))))))))...........(((.....------...)))...................................... ( -15.70) >DroSec_CAF1 57350 92 - 1 CUC------ACACACACACGAG---UAGUAUAUGUGUGCACCAACACAAGCGAACUU------CAACGCAAUUGUCAUCACCAUAAAAAUAACAUAAAACUUUUUUA ...------..........(((---(..(((.(((((......))))).(((.....------...)))........................)))..))))..... ( -10.70) >DroSim_CAF1 44870 93 - 1 CUC------ACACACACACAAA--CGAGUAUGUGUGUGCACCAACACAAGCGAACUU------CAACGCAAUUGUCAUCACCAUAAAAAUAACAUAAAACUUUUUUA ..(------(((((((.((...--...)).))))))))...........(((.....------...)))...................................... ( -15.30) >DroYak_CAF1 60682 101 - 1 CCC------GCACACACACACAUAUACGUGUGUGUGCCCACCAACACAAGCGAACGCGAACGCAAACGCAAUUGUCAUCACCAUAAAAAUAACAUAAAACUUUUUUA ...------((((((((((........))))))))))............(((...((....))...)))...................................... ( -23.10) >consensus CUC______ACACACACACAAA__CGAGUAUGUGUGUGCACCAACACAAGCGAACUU______CAACGCAAUUGUCAUCACCAUAAAAAUAACAUAAAACUUUUUUA .........(((((((.((........)).)))))))............(((..............)))...................................... (-11.19 = -11.07 + -0.12)

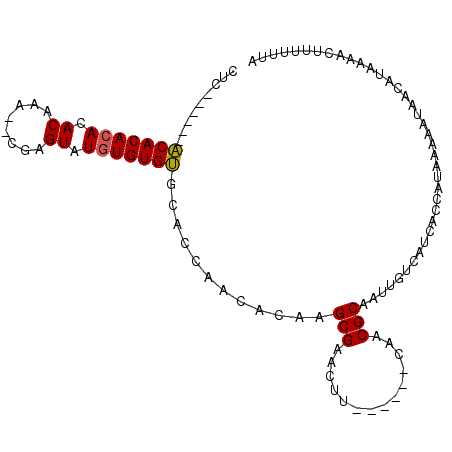
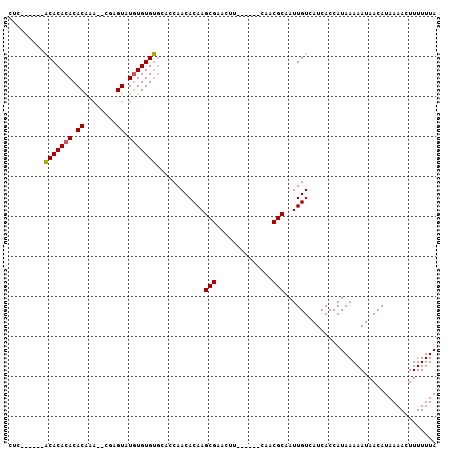
| Location | 6,489,655 – 6,489,750 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -25.05 |
| Energy contribution | -24.93 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.81 |
| Structure conservation index | 0.83 |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.994695 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6489655 95 + 20766785 CGUUG------AAGUUCGCUUGUGUUGGUGCACACACAUACGCG--UGUGUGUGUGUGUGUGAGAGAGUAUUAUUGUUGCUUACUCAACAUGCAUUUAUGCCC .....------(((....)))((((..((((((((((((((((.--...)))))))))))((((.(((((.......))))).))))...)))))..)))).. ( -32.30) >DroSec_CAF1 57389 88 + 1 CGUUG------AAGUUCGCUUGUGUUGGUGCACACAUAUACUA---CUCGUGUGUGUGU------GAGUAUUAUUGUUGCUUACUCAACAUGCAUUUAUGCCC .((((------(.((..((......((((((.(((((((((..---.....))))))))------).)))))).....))..)))))))..(((....))).. ( -26.40) >DroSim_CAF1 44909 89 + 1 CGUUG------AAGUUCGCUUGUGUUGGUGCACACACAUACUCG--UUUGUGUGUGUGU------GAGUAUUAUUGUUGCUUACUCAACAUGCAUUUAUGCCC .((((------(.((..((......((((((.(((((((((...--.....))))))))------).)))))).....))..)))))))..(((....))).. ( -28.60) >DroYak_CAF1 60721 97 + 1 CGUUUGCGUUCGCGUUCGCUUGUGUUGGUGGGCACACACACGUAUAUGUGUGUGUGUGC------GGGUGUUAUUGUUGCUUACUCAACAUGCAUUUAUGCCC .....(((..((((((((((......)))))))(((((((((......)))))))))))------)..)))...(((((......))))).(((....))).. ( -33.30) >consensus CGUUG______AAGUUCGCUUGUGUUGGUGCACACACAUACGCG__UGUGUGUGUGUGU______GAGUAUUAUUGUUGCUUACUCAACAUGCAUUUAUGCCC .................((..((((((((((((((((((((......))))))))))))......(((((.......))))))).)))))))).......... (-25.05 = -24.93 + -0.12)

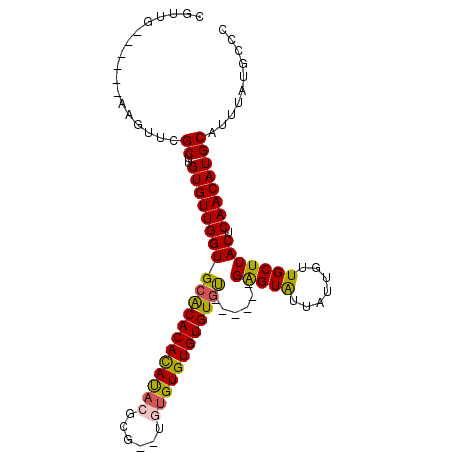

| Location | 6,489,655 – 6,489,750 |
|---|---|
| Length | 95 |
| Sequences | 4 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 83.33 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -16.75 |
| Energy contribution | -16.62 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.63 |
| Structure conservation index | 0.74 |
| SVM decision value | 1.91 |
| SVM RNA-class probability | 0.982337 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6489655 95 - 20766785 GGGCAUAAAUGCAUGUUGAGUAAGCAACAAUAAUACUCUCUCACACACACACACACA--CGCGUAUGUGUGUGCACCAACACAAGCGAACUU------CAACG (((((....))).(((((......)))))..............((((((((.((...--...)).))))))))..))...............------..... ( -20.00) >DroSec_CAF1 57389 88 - 1 GGGCAUAAAUGCAUGUUGAGUAAGCAACAAUAAUACUC------ACACACACACGAG---UAGUAUAUGUGUGCACCAACACAAGCGAACUU------CAACG ..(((....)))..(((((((..((....(((.(((((------..........)))---)).))).(((((......))))).))..)).)------)))). ( -20.80) >DroSim_CAF1 44909 89 - 1 GGGCAUAAAUGCAUGUUGAGUAAGCAACAAUAAUACUC------ACACACACACAAA--CGAGUAUGUGUGUGCACCAACACAAGCGAACUU------CAACG ..(((....)))..(((((((..((............(------(((((((.((...--...)).))))))))...........))..)).)------)))). ( -21.80) >DroYak_CAF1 60721 97 - 1 GGGCAUAAAUGCAUGUUGAGUAAGCAACAAUAACACCC------GCACACACACACAUAUACGUGUGUGUGCCCACCAACACAAGCGAACGCGAACGCAAACG (((((....))).(((((......))))).......))------((((((((((........))))))))))............(((........)))..... ( -28.50) >consensus GGGCAUAAAUGCAUGUUGAGUAAGCAACAAUAAUACUC______ACACACACACAAA__CGAGUAUGUGUGUGCACCAACACAAGCGAACUU______CAACG (((((....))).(((((......)))))...............(((((((.((........)).)))))))...)).......................... (-16.75 = -16.62 + -0.12)

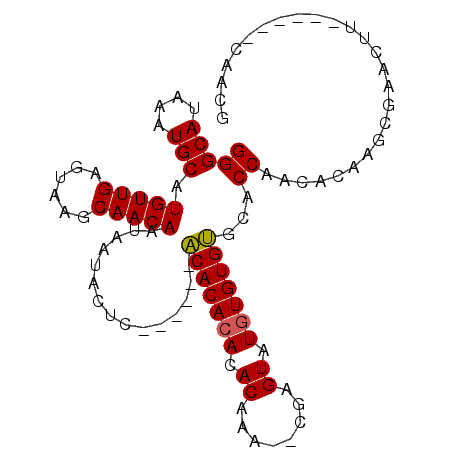
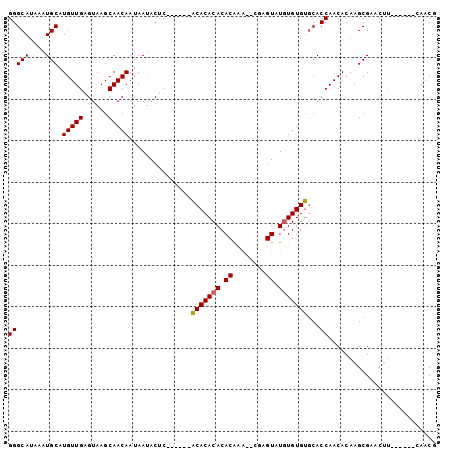
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:45 2006