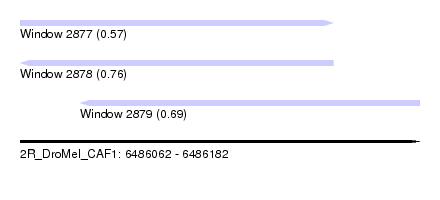
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,486,062 – 6,486,182 |
| Length | 120 |
| Max. P | 0.755170 |

| Location | 6,486,062 – 6,486,156 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -25.07 |
| Energy contribution | -26.90 |
| Covariance contribution | 1.83 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.567455 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6486062 94 + 20766785 AGGCUGCGAAGGCUGGGAAGCCAGG--AUCUGCGUCCUGCAGACAAACUUCCGCUGGAGGAAAUUUUAUGCAGGGCACGGCAAAAGGAUGCGCAGC ..((((((..((((....))))...--..(((.((((((((...(((.((((......)))).)))..)))))))).)))..........)))))) ( -38.90) >DroSec_CAF1 53941 94 + 1 AGGUUGCGCAGGCUGGGAAGCCAGG--AUCUGCGUCCUGCAGACAAACUUCCGCUGGAGGAAAUUUUAUGCAGGGCACGGCAAAAGGAUGCGCAGC ..((((((((((((....))))...--..(((.((((((((...(((.((((......)))).)))..)))))))).)))........)))))))) ( -41.30) >DroSim_CAF1 41495 94 + 1 AGGUUGCGCAGGCUGGGAAGCCAGG--AUCUGCGUCCUGCAGACAAACUUCCGCUGGAGGAAAUUUUAUGCAGGGCACGGCAAAAGGAUGCGCAGC ..((((((((((((....))))...--..(((.((((((((...(((.((((......)))).)))..)))))))).)))........)))))))) ( -41.30) >DroEre_CAF1 52196 81 + 1 -------------UGGGAAGCCAGG--AUCUGCGUCCUGCAGACAAACUUCCGCUGCUGGAAAUUUUAUGCAGGGCACGGCAAAAGGAUGCGCAGC -------------(((....)))..--..((((((((((((...(((.(((((....))))).)))..))))))))...(((......))))))). ( -29.70) >DroAna_CAF1 53265 84 + 1 -------GAGGGCUC---UGCCAGG--AUCUGCGUCCUGCAGACAAACUUCCGCUGGAGGAAAUUUUAUGCAGGGCACGGCAAAAGGAUGCGCAGC -------...(((..---.)))...--..((((((((((((...(((.((((......)))).)))..))))))))...(((......))))))). ( -30.20) >DroPer_CAF1 50912 92 + 1 AGG--GGGCAGG--GGGCGGCGAGGGGUACCCCGGGCUGCAGACAAACUUCCGCUAAAGGAAAUUUUAUGCAGGGCACGGCCAAAGGAUGCGCAGC ...--(.(((..--((.((((..(((....)))...(((((...(((.((((......)))).)))..))))).)).)).))......))).)... ( -31.80) >consensus AGG__GCGCAGGCUGGGAAGCCAGG__AUCUGCGUCCUGCAGACAAACUUCCGCUGGAGGAAAUUUUAUGCAGGGCACGGCAAAAGGAUGCGCAGC ............((((....)))).....((((((((((((...(((.((((......)))).)))..))))))))...(((......))))))). (-25.07 = -26.90 + 1.83)

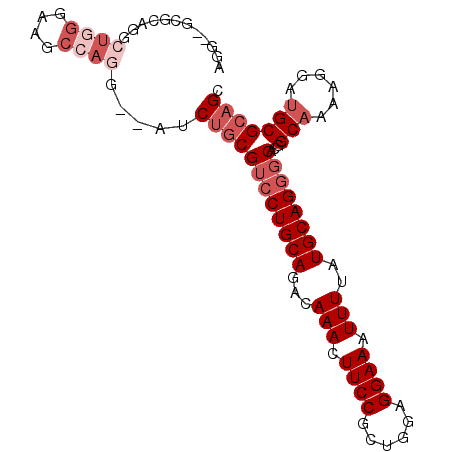
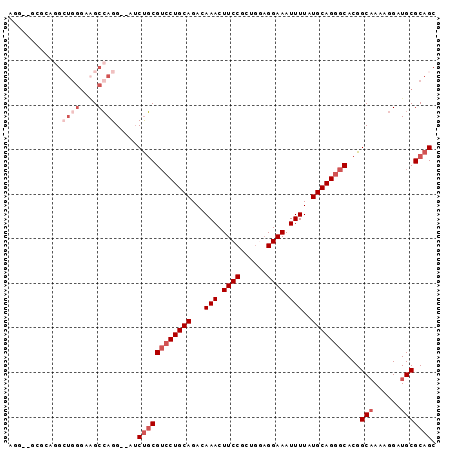
| Location | 6,486,062 – 6,486,156 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | reverse |
| Mean pairwise identity | 84.87 |
| Mean single sequence MFE | -33.30 |
| Consensus MFE | -24.16 |
| Energy contribution | -24.52 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.755170 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6486062 94 - 20766785 GCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUCCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGCUUCCCAGCCUUCGCAGCCU ((((((((......))).(((.((((((..((((((((......))))))))...)))))).))).((.--...((((....)))).))))))).. ( -38.40) >DroSec_CAF1 53941 94 - 1 GCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUCCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGCUUCCCAGCCUGCGCAACCU ..((((((..........(((.((((((..((((((((......))))))))...)))))).)))....--...((((....)))))))))).... ( -36.20) >DroSim_CAF1 41495 94 - 1 GCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUCCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGCUUCCCAGCCUGCGCAACCU ..((((((..........(((.((((((..((((((((......))))))))...)))))).)))....--...((((....)))))))))).... ( -36.20) >DroEre_CAF1 52196 81 - 1 GCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCAGCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGCUUCCCA------------- ((((((((......))).((((...))))..........)))))(((((((.((((((.....))))))--...)))))))..------------- ( -30.70) >DroAna_CAF1 53265 84 - 1 GCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUCCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGCA---GAGCCCUC------- .(((((((......))).(((.((((((..((((((((......))))))))...)))))).)))....--....)))---).......------- ( -30.60) >DroPer_CAF1 50912 92 - 1 GCUGCGCAUCCUUUGGCCGUGCCCUGCAUAAAAUUUCCUUUAGCGGAAGUUUGUCUGCAGCCCGGGGUACCCCUCGCCGCCC--CCUGCCC--CCU .....(((......(((.(((..(((((..((((((((......))))))))...)))))...(((....))).))).))).--..)))..--... ( -27.70) >consensus GCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUCCAGCGGAAGUUUGUCUGCAGGACGCAGAU__CCUGGCUUCCCAGCCUGCGC__CCU .(((((((......)))...(.((((((..((((((((......))))))))...)))))).)))))............................. (-24.16 = -24.52 + 0.36)
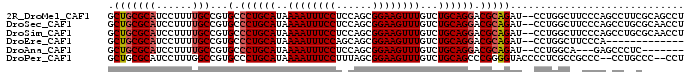
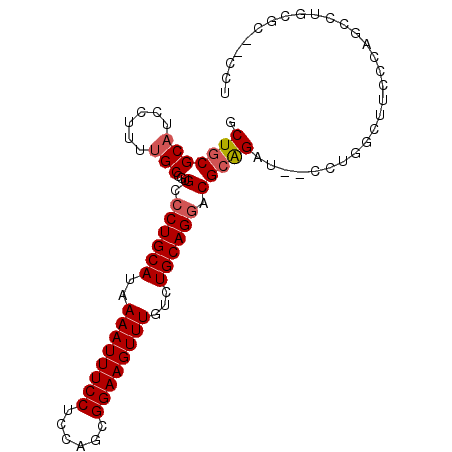
| Location | 6,486,080 – 6,486,182 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 83.91 |
| Mean single sequence MFE | -32.35 |
| Consensus MFE | -25.72 |
| Energy contribution | -24.89 |
| Covariance contribution | -0.83 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.84 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.33 |
| SVM RNA-class probability | 0.690470 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6486080 102 - 20766785 ---ACAUGG-C----------AUCCAUGUAUUAUUUAAGCGCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUCCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGC--U ---((((((-.----------..))))))...........((((.(((......))))(((.((((((..((((((((......))))))))...)))))).)))....--...)))--. ( -32.80) >DroPse_CAF1 51245 104 - 1 ---ACAUGG-C----------AUCCAUGUAUUAUUUAAGCGCUGCGCAUCCUUUGGCCGUGCCCUGCAUAAAAUUUCCUUUAGCGGAAGUUUGUCUGCAGCCCGGGGUACCCCUCGC--C ---((((((-.----------..)))))).........((((.((.((.....)))).)))).(((((..((((((((......))))))))...)))))...(((....)))....--. ( -33.30) >DroEre_CAF1 52201 102 - 1 ---ACAUGG-C----------AUCCAUGUAUUAUUUAAGCGCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCAGCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGC--U ---((((((-.----------..))))))...........((((.(((......))))(((.((((((..((((((((......))))))))...)))))).)))....--...)))--. ( -32.90) >DroWil_CAF1 68095 118 - 1 AGGAUAUGGGCAACUGUCCGGCUCCAUGUAUUAUUUAAGCAUUGCGCAUCCUUUUGCCGUGCCUUGCAUAAAAUUUCAUUCUGCGGAAGUUUGUCUGCAGCUCCGUCAC--CGUCACCAU .(((..(((((....)))))..)))........((((.(((..((((...........))))..))).))))........(((((((......))))))).........--......... ( -28.50) >DroAna_CAF1 53273 102 - 1 ---ACAUGG-C----------AUCCAUGUAUUAUUUAAGCGCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUCCAGCGGAAGUUUGUCUGCAGGACGCAGAU--CCUGGC--A ---((((((-.----------..))))))...........((((.(((......))))(((.((((((..((((((((......))))))))...)))))).)))....--...)))--. ( -33.30) >DroPer_CAF1 50926 104 - 1 ---ACAUGG-C----------AUCCAUGUAUUAUUUAAGCGCUGCGCAUCCUUUGGCCGUGCCCUGCAUAAAAUUUCCUUUAGCGGAAGUUUGUCUGCAGCCCGGGGUACCCCUCGC--C ---((((((-.----------..)))))).........((((.((.((.....)))).)))).(((((..((((((((......))))))))...)))))...(((....)))....--. ( -33.30) >consensus ___ACAUGG_C__________AUCCAUGUAUUAUUUAAGCGCUGCGCAUCCUUUUGCCGUGCCCUGCAUAAAAUUUCCUUCAGCGGAAGUUUGUCUGCAGCACGCAGAU__CCUCGC__U ...((((((..............)))))).........((((.(((........))).)))).(((((..((((((((......))))))))...)))))....(((.....)))..... (-25.72 = -24.89 + -0.83)
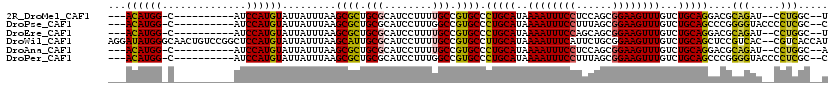
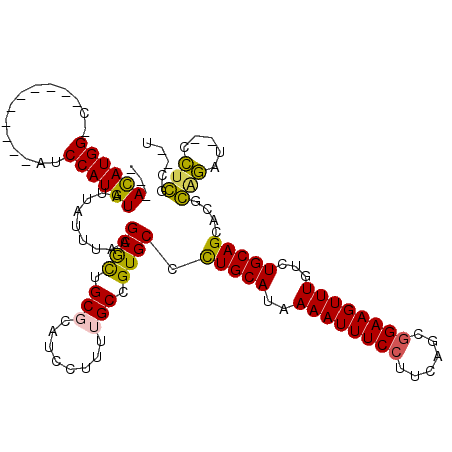
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:42 2006