
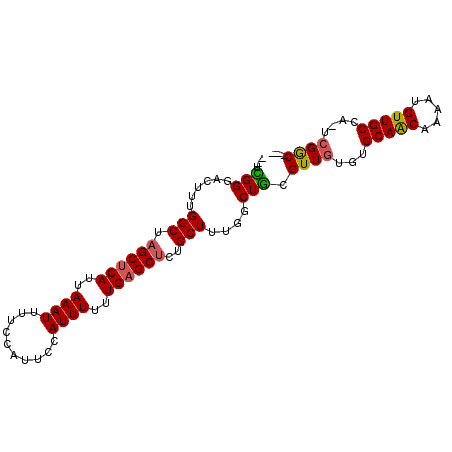
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,477,276 – 6,477,441 |
| Length | 165 |
| Max. P | 0.815367 |

| Location | 6,477,276 – 6,477,369 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 87.03 |
| Mean single sequence MFE | -23.28 |
| Consensus MFE | -16.94 |
| Energy contribution | -16.74 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.660731 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
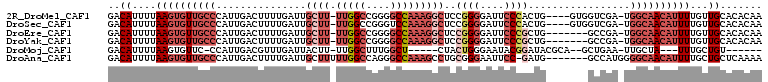
>2R_DroMel_CAF1 6477276 93 + 20766785 --UCGGGACUUUGGCUAGCUCAUUAAAUUUUCCAUUCCAUUUUUUGAGCUCUGCUUUGGCUGCGUUGUGUGCAACAAAAUGUUGCCA-UCGACCAC --...((.((..(((.((((((..((((..........))))..))))))..)))..))))(.((((((.(((((.....)))))))-.))))).. ( -25.40) >DroSec_CAF1 45408 93 + 1 --UCGGGACUUUGGCUAGCUCAUUAAAUUUUCCAUUCCAUUUUUUGAGCUCUGCUUUGGCUGCGUUGUGUGCAACAAAAUGUUGCCA-UCGACCAC --...((.((..(((.((((((..((((..........))))..))))))..)))..))))(.((((((.(((((.....)))))))-.))))).. ( -25.40) >DroEre_CAF1 43574 90 + 1 --UCGGGACUUUGGCUAGCUCAUUAAAUUUUCCAUUCCAUUUUUUGAGCUCUGCUUUGACUGCGUUGUGUGCAACAAAAUGUUGCCA-UCGGC--- --.(((......(((.((((((..((((..........))))..))))))..)))....))).((((((.(((((.....)))))))-.))))--- ( -22.30) >DroYak_CAF1 48101 90 + 1 --UUGGGACUUUGGCUAGCUCAUUAAAUUUUCCAUUCCAUUUUUUGAGCUCUGCUUUGGCUGCGUUGUGUGCAACAAAAUGUUGCCA-UCGGC--- --...((.((..(((.((((((..((((..........))))..))))))..)))..))))..((((((.(((((.....)))))))-.))))--- ( -24.00) >DroAna_CAF1 45382 86 + 1 CUUUGGGACUCUGGCUAGCUCAUUAAAUUUUCC-----AUUUUUUGUGCGAUGCUUU--CUAUGUUUUGAGCAGCAAAAUGUUGCCCCAUGGC--- ((.((((.....(((..((.((..((((.....-----))))..)).))...)))..--...........(((((.....))))))))).)).--- ( -19.30) >consensus __UCGGGACUUUGGCUAGCUCAUUAAAUUUUCCAUUCCAUUUUUUGAGCUCUGCUUUGGCUGCGUUGUGUGCAACAAAAUGUUGCCA_UCGGC___ ...(((......(((.((((((..((((..........))))..))))))..)))....))).((((...(((((.....)))))....))))... (-16.94 = -16.74 + -0.20)


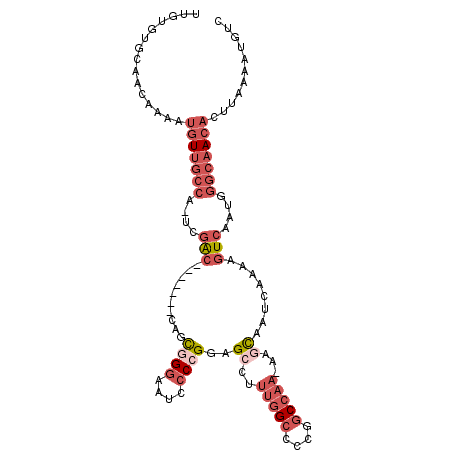
| Location | 6,477,338 – 6,477,441 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -28.17 |
| Consensus MFE | -15.69 |
| Energy contribution | -17.92 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.94 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.586745 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6477338 103 + 20766785 UUGUGUGCAACAAAAUGUUGCCA-UCGACCAC----CAGUGGGAAUCCCCGGAGCCUUUGGCCCCGGCCAA-AAGCAAUCAAAAGUCAAUGGGCAACACUUAAAAUGUC ((((.....))))..(((((((.-..(((...----...((((....))))..((.((((((....)))))-).))........)))....)))))))........... ( -31.50) >DroSec_CAF1 45470 103 + 1 UUGUGUGCAACAAAAUGUUGCCA-UCGACCAC----CAGUGGGAAUCCCCGGAGCCUUUGGACCCGGCCAA-AAGCAAUCAAAAGUCAAUGGGCAACACUUAAAAUGUC ((((.....))))..(((((((.-..(((...----...((((....))))..((.(((((......))))-).))........)))....)))))))........... ( -26.70) >DroEre_CAF1 43636 100 + 1 UUGUGUGCAACAAAAUGUUGCCA-UCGGC-------CAGCGGGAAUCCCCGGAGCCUUUGGCCCCGGCCAA-AAGCAAUCAAAAGUCAAUGGGCAACACUUAAAAUGUC ..(((((((((.....)))))..-...((-------(..((((....))))..((.((((((....)))))-).))...............))).)))).......... ( -33.20) >DroYak_CAF1 48163 100 + 1 UUGUGUGCAACAAAAUGUUGCCA-UCGGC-------CAGCGGGAAUCCCCGGAGCCUUUGGCCCCGGCCAA-AAGCAAUCAAAAGUCAAUGGGCAACACUUAAAAUGUC ..(((((((((.....)))))..-...((-------(..((((....))))..((.((((((....)))))-).))...............))).)))).......... ( -33.20) >DroMoj_CAF1 38983 90 + 1 ------ACAGCAAA---UAGCAA-UUCAGC--UGCGUAUCCGUAUUCCCAGUAG-----AGCCAAAGCCAA-AAGUAAUCAAACGUCAAUGG-GAACACUUAAAAUGUC ------...(((..---((((..-....))--)).......((.((((((...(-----(((....))...-..((......)).))..)))-))).))......))). ( -12.80) >DroAna_CAF1 45439 101 + 1 UUUUGAGCAGCAAAAUGUUGCCCCAUGGC-------CAUC-GGAAUUCCCGCAGGCUUUGGCCCUGGCCAAAAAGCAAUCAAAAGUCAAUGGGCAACACUUAAAAUGUC (((((((........((((((((..((((-------...(-((.....)))...((((((((....))))..))))........))))..))))))))))))))).... ( -31.60) >consensus UUGUGUGCAACAAAAUGUUGCCA_UCGAC_______CAGCGGGAAUCCCCGGAGCCUUUGGCCCCGGCCAA_AAGCAAUCAAAAGUCAAUGGGCAACACUUAAAAUGUC ...............(((((((....(((..........((((....))))..((..(((((....)))))...))........)))....)))))))........... (-15.69 = -17.92 + 2.22)
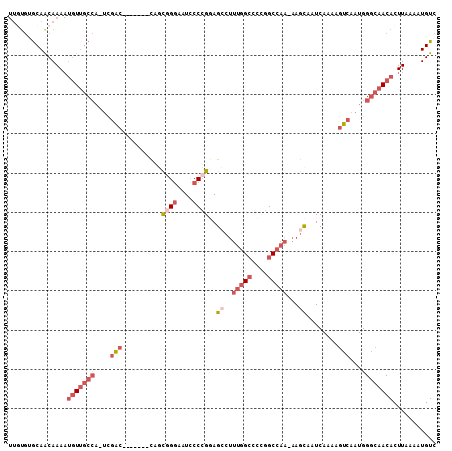
| Location | 6,477,338 – 6,477,441 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -35.53 |
| Consensus MFE | -17.04 |
| Energy contribution | -19.85 |
| Covariance contribution | 2.81 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.75 |
| Structure conservation index | 0.48 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.815367 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6477338 103 - 20766785 GACAUUUUAAGUGUUGCCCAUUGACUUUUGAUUGCUU-UUGGCCGGGGCCAAAGGCUCCGGGGAUUCCCACUG----GUGGUCGA-UGGCAACAUUUUGUUGCACACAA ((((....((((((((((.(((((((.......((((-(((((....))))))))).((((((....)).)))----).))))))-)))))))))))))))........ ( -43.50) >DroSec_CAF1 45470 103 - 1 GACAUUUUAAGUGUUGCCCAUUGACUUUUGAUUGCUU-UUGGCCGGGUCCAAAGGCUCCGGGGAUUCCCACUG----GUGGUCGA-UGGCAACAUUUUGUUGCACACAA ((((....((((((((((.(((((((.......((((-((((......)))))))).((((((....)).)))----).))))))-)))))))))))))))........ ( -37.90) >DroEre_CAF1 43636 100 - 1 GACAUUUUAAGUGUUGCCCAUUGACUUUUGAUUGCUU-UUGGCCGGGGCCAAAGGCUCCGGGGAUUCCCGCUG-------GCCGA-UGGCAACAUUUUGUUGCACACAA ..........((((.((.....(((...((.(((((.-((((((((((((...))))))(((....)))...)-------)))))-.)))))))....))))))))).. ( -38.30) >DroYak_CAF1 48163 100 - 1 GACAUUUUAAGUGUUGCCCAUUGACUUUUGAUUGCUU-UUGGCCGGGGCCAAAGGCUCCGGGGAUUCCCGCUG-------GCCGA-UGGCAACAUUUUGUUGCACACAA ..........((((.((.....(((...((.(((((.-((((((((((((...))))))(((....)))...)-------)))))-.)))))))....))))))))).. ( -38.30) >DroMoj_CAF1 38983 90 - 1 GACAUUUUAAGUGUUC-CCAUUGACGUUUGAUUACUU-UUGGCUUUGGCU-----CUACUGGGAAUACGGAUACGCA--GCUGAA-UUGCUA---UUUGCUGU------ ..........((((((-(((.....((......))..-..(((....)))-----....)))))))))......(((--((.(((-(....)---))))))))------ ( -20.70) >DroAna_CAF1 45439 101 - 1 GACAUUUUAAGUGUUGCCCAUUGACUUUUGAUUGCUUUUUGGCCAGGGCCAAAGCCUGCGGGAAUUCC-GAUG-------GCCAUGGGGCAACAUUUUGCUGCUCAAAA ((((....(((((((((((..((..............((((((....))))))(((..(((.....))-)..)-------))))..)))))))))))...)).)).... ( -34.50) >consensus GACAUUUUAAGUGUUGCCCAUUGACUUUUGAUUGCUU_UUGGCCGGGGCCAAAGGCUCCGGGGAUUCCCGCUG_______GCCGA_UGGCAACAUUUUGUUGCACACAA ..((....((((((((((...............((((.(((((....)))))))))..((((....)))).................))))))))))...))....... (-17.04 = -19.85 + 2.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:39 2006