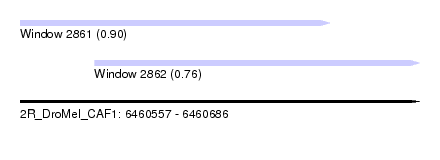
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,460,557 – 6,460,686 |
| Length | 129 |
| Max. P | 0.897363 |

| Location | 6,460,557 – 6,460,657 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 79.97 |
| Mean single sequence MFE | -27.92 |
| Consensus MFE | -16.89 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.00 |
| SVM RNA-class probability | 0.897363 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
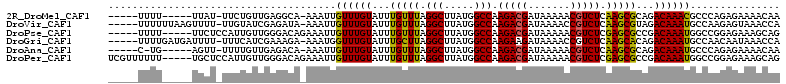
>2R_DroMel_CAF1 6460557 100 + 20766785 -----UUUU-----UUAU-UUCUGUUGAGGCA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCAAGCGCAGACAAACGCCCAGAGAAAACAA -----....-----...(-((((.(((.(((.-...(((((((..((((....(((.....))).(((((.......)))))))))..)))))))..))))))))))).... ( -28.80) >DroVir_CAF1 26106 105 + 1 -----UUUUUUAAGUUUU-UUGUAUCGAGAUA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAACCGUCUCAAGCGUAGACAAAUGCCAAGAGUAAACCA -----........(((((-((((......)))-))(((.((.(((((((((((.((((.......(((((.......))))).)))).))))))))))).)).))))))).. ( -22.10) >DroPse_CAF1 37471 102 + 1 -----UUUU-----UUCUCCAUUGUUGGGACAGAAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCGAGCGCCGACAAAUGGCCGGAGAAAGCAG -----..((-----((((((....((((((((....).((((....((((((.(((.....))).))))))...)))))))))))(.((((.....)))))))))))))... ( -32.10) >DroGri_CAF1 17747 105 + 1 -----UUUUGAUGAUUUU-UUUCAUCGAAAGA-AAAUGGUUUGUAUUUGCUUAGGCUUAUGGCCAAGAAGAUAAAACCGUCUCAAGCACAGACAAAUGCCAACAAUAAACCA -----(((((((((....-..)))))))))..-...((((((((...(((((.(((.....)))..((.(((......))))))))))...))))..))))........... ( -26.60) >DroAna_CAF1 37629 99 + 1 -----C-UG-----AGUU-UUUUGUUGAGACA-AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAA -----.-..-----.(((-((((.(((((((.-.....((((....((((((.(((.....))).))))))...)))))))))))(.(((......))).)..))))))).. ( -25.80) >DroPer_CAF1 37140 107 + 1 UCGUUUUUU-----UGCUCCAUUGUUGGGACAGAAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCGAGCGCCGACAAAUGGCCGGAGAAAGCAG ..(((((((-----(...(((((((((((...((...(((((....((((((.(((.....))).))))))...)))))..))...).))))).)))))...)))))))).. ( -32.10) >consensus _____UUUU_____UUUU_UUUUGUUGAGACA_AAAUUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAA ......................................((((((...(((((.(((.....))).(((((.......))))).)))))...))))))............... (-16.89 = -16.42 + -0.47)

| Location | 6,460,581 – 6,460,686 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 78.43 |
| Mean single sequence MFE | -27.03 |
| Consensus MFE | -18.24 |
| Energy contribution | -18.22 |
| Covariance contribution | -0.03 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.764507 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6460581 105 + 20766785 UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCAAGCGCAGACAAACGCCCAGAGAAAACAA-GUG--AAUGACG-CUGACU-UGGCGCAAU-CGAA (((((((..((((....(((.....))).(((((.......)))))))))..))))))).((((..(((......(-(((--.....))-))..))-)))))....-.... ( -27.20) >DroVir_CAF1 26135 104 + 1 UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAACCGUCUCAAGCGUAGACAAAUGCCAAGAGUAAACCAAGAA--AAUU----UGUAAA-UAUAAUAUAAUGAA ((.((.(((((((((((.((((.......(((((.......))))).)))).))))))))))).)).))...........--....----......-.............. ( -20.20) >DroPse_CAF1 37497 107 + 1 UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCGAGCGCCGACAAAUGGCCGGAGAAAGCAG-GCG--GAUGACGGCCGACUUUAGAGCCAU-CGAA ..((((....((((((.(((.....))).))))))...))))...(((((.((((.....)))))((..((((..(-((.--(....).)))..))))....)).)-))). ( -31.00) >DroMoj_CAF1 32079 106 + 1 UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAACCGUCUCAAGCGCAGACAAAUGCCAACAAUAAGCCAAGAACAAAUU----UAUGCA-CUUAAUACACUGAA .(((....((((((((.(((((((.((..(((((.......)))))...))(((......))).....)))))))..)))))))).----...)))-.............. ( -25.80) >DroAna_CAF1 37652 105 + 1 UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAA-CUG--ACUGAGC-CUGACU-UGGCACAAU-CGAA (((((((..((((....(((.....))).(((((.......)))))))))..))))))).................-.((--(.((.((-(.....-.))).)).)-)).. ( -27.00) >DroPer_CAF1 37171 107 + 1 UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCGAGCGCCGACAAAUGGCCGGAGAAAGCAG-GCG--GAUGACGGCCGACUUUAGAGCCAU-CGAA ..((((....((((((.(((.....))).))))))...))))...(((((.((((.....)))))((..((((..(-((.--(....).)))..))))....)).)-))). ( -31.00) >consensus UUGUUUGUAUUUGUUUAGGCUUAUGGCCAAGACGAUAAAAACGUCUCAAGCGCAGACAAAUGCCCAGAGAAAACAA_GAG__AAUGACG_CCGACU_UAGAACAAU_CGAA (((((((..((((....(((.....))).(((((.......)))))))))..))))))).................................................... (-18.24 = -18.22 + -0.03)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:26 2006