
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,458,414 – 6,458,571 |
| Length | 157 |
| Max. P | 0.999818 |

| Location | 6,458,414 – 6,458,505 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -17.52 |
| Energy contribution | -18.52 |
| Covariance contribution | 1.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.90 |
| SVM decision value | 2.68 |
| SVM RNA-class probability | 0.996329 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6458414 91 + 20766785 GAAUGGCGCCCACCCUUUUGUACCCAUACUGCCCAUCUCUCUCGGCAUUGUUCUACCCAUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCA ..((((.(..((......))..)))))..((((..........))))............(((((((.....)))))))..(((....))). ( -15.00) >DroSec_CAF1 34048 90 + 1 GAAUGGCGCCCUCCCUUUUGUGCCCAUUCUACCCGUCUC-CUCGGCAUUGUUCUACCCGUCCCUUUCUCAUAAAGAGACUGCAGUUAUGCA ((((((.((.(........).))))))))...(((....-..))).............(((.((((.....)))).))).(((....))). ( -19.10) >DroSim_CAF1 25410 91 + 1 GAAUGGCGCCCUCCCUUUUGUGCCCAUUCUACCCGUCUCUCUCGGCAUUGUUCUACCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCA ((((((.((.(........).))))))))...(((.......))).............((((((((.....)))))))).(((....))). ( -23.40) >DroEre_CAF1 32368 90 + 1 GAAUGGCGCCCUC-CUUUUGUGCCCACUCUACCCAUCUCUCUCGGCAUUGUUCUACCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCA (((((((((....-.....)))))........((.........))....)))).....((((((((.....)))))))).(((....))). ( -18.20) >DroYak_CAF1 36010 91 + 1 GAAUGGCGCCCUCCCUUUUGUGCCCAUUCUACCCAUCUCUCUCGGCAUUGUUCUACCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCA ((((((.((.(........).))))))))...............((((....((.(..((((((((.....)))))))).).))..)))). ( -22.00) >consensus GAAUGGCGCCCUCCCUUUUGUGCCCAUUCUACCCAUCUCUCUCGGCAUUGUUCUACCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCA ((((((.((.(........).))))))))...............((((....((..(.((((((((.....)))))))).).))..)))). (-17.52 = -18.52 + 1.00)

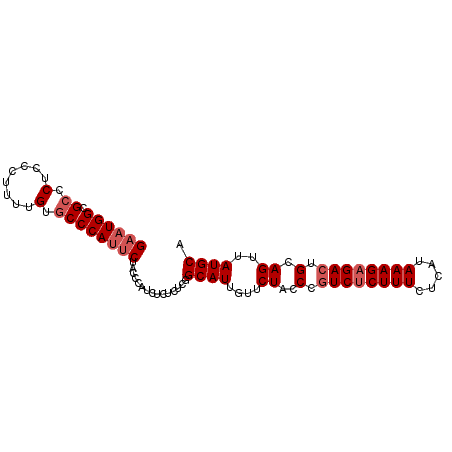
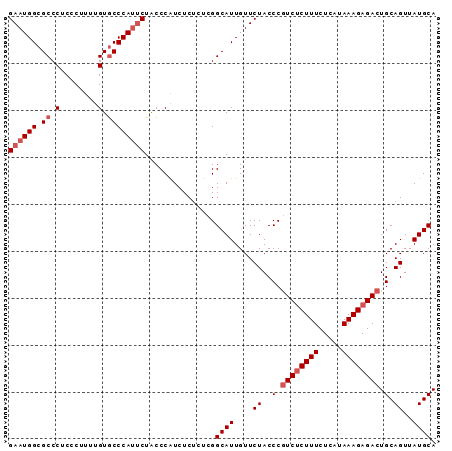
| Location | 6,458,414 – 6,458,505 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | reverse |
| Mean pairwise identity | 95.38 |
| Mean single sequence MFE | -26.82 |
| Consensus MFE | -25.28 |
| Energy contribution | -24.68 |
| Covariance contribution | -0.60 |
| Combinations/Pair | 1.20 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.16 |
| SVM RNA-class probability | 0.999818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6458414 91 - 20766785 UGCAUAACUGCAGUCUCUUUAUGAGAAAGAGAUGGGUAGAACAAUGCCGAGAGAGAUGGGCAGUAUGGGUACAAAAGGGUGGGCGCCAUUC ..((((.((((.((((((((.....))))))))..)))).....((((.(......).)))).)))).........((((((...)))))) ( -24.50) >DroSec_CAF1 34048 90 - 1 UGCAUAACUGCAGUCUCUUUAUGAGAAAGGGACGGGUAGAACAAUGCCGAG-GAGACGGGUAGAAUGGGCACAAAAGGGAGGGCGCCAUUC .......((((.((((((((.....))))))))..)))).....((((..(-....).))))((((((((.(........).)).)))))) ( -28.70) >DroSim_CAF1 25410 91 - 1 UGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGACGGGUAGAAUGGGCACAAAAGGGAGGGCGCCAUUC (((....((((.((((((((.....))))))))..)))).......(((.......))))))((((((((.(........).)).)))))) ( -26.50) >DroEre_CAF1 32368 90 - 1 UGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGAUGGGUAGAGUGGGCACAAAAG-GAGGGCGCCAUUC .......((((.((((((((.....))))))))..)))).....((((.(......).))))((((((((.(.....-...))).)))))) ( -27.10) >DroYak_CAF1 36010 91 - 1 UGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGAUGGGUAGAAUGGGCACAAAAGGGAGGGCGCCAUUC .......((((.((((((((.....))))))))..)))).....((((.(......).))))((((((((.(........).)).)))))) ( -27.30) >consensus UGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGAUGGGUAGAAUGGGCACAAAAGGGAGGGCGCCAUUC .......((((.((((((((.....))))))))..)))).....((((..........))))((((((((.(........).)).)))))) (-25.28 = -24.68 + -0.60)

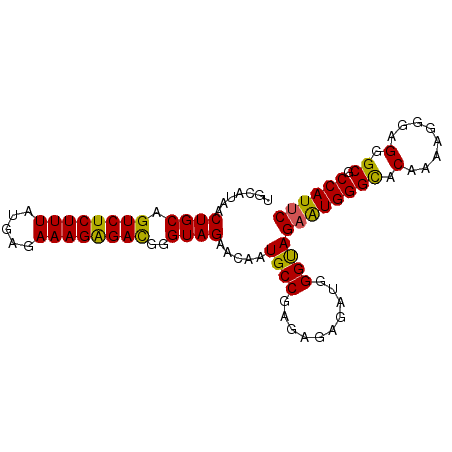
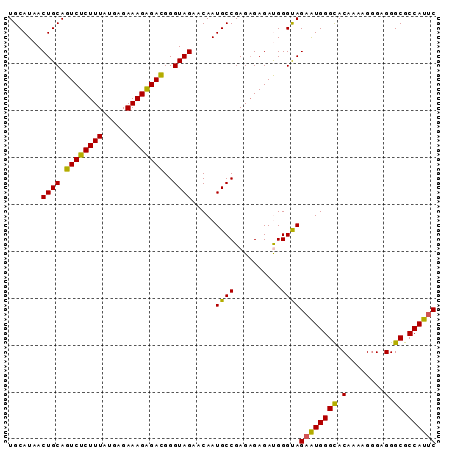
| Location | 6,458,439 – 6,458,540 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 88.12 |
| Mean single sequence MFE | -28.20 |
| Consensus MFE | -22.84 |
| Energy contribution | -23.20 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.16 |
| Structure conservation index | 0.81 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.680060 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6458439 101 - 20766785 GCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGAUGGGUAGAACAAUGCCGAGAGAGAUGGGCAGUAU ((.((....))..(((((.....-----)))))...)).((((....((((.((((((((.....))))))))..)))).....((((.(......).)))))))) ( -30.20) >DroSec_CAF1 34073 100 - 1 GCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGGGACGGGUAGAACAAUGCCGAG-GAGACGGGUAGAAU (((((....))..(((((.....-----))))).......)))....((((.((((((((.....))))))))..)))).....((((..(-....).)))).... ( -30.30) >DroSim_CAF1 25435 101 - 1 GCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGACGGGUAGAAU (((((....))..(((((.....-----))))).......)))....((((.((((((((.....)))))))).((((......))))...........))))... ( -28.40) >DroEre_CAF1 32392 101 - 1 GCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGAUGGGUAGAGU (((((....))..(((((.....-----))))).......)))....((((.((((((((.....))))))))..)))).((..((((.(......).))))..)) ( -29.50) >DroYak_CAF1 36035 101 - 1 GCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGAUGGGUAGAAU (((((....))..(((((.....-----))))).......)))....((((.((((((((.....))))))))..)))).....((((.(......).)))).... ( -28.90) >DroAna_CAF1 35576 94 - 1 GCAGAGCAAUCAAGCGCGAAAUCUCUGCUGUGCACAGCAAUGCAUAACUGCAGUCUCCGUGUGAGACAAAAACAG---------U---GAAAGAGAAGGGACUAGU ((.((....))..))......(((((((((....))))..((((....))))(((((.....)))))........---------.---...))))).......... ( -21.90) >consensus GCAGAGCAAUCAAGCGCGAAAUC_____UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGGUAGAACAAUGCCGAGAGAGAUGGGUAGAAU ((((.((......)).............((((((......)))))).)))).((((((((.....)))))))).((((......)))).................. (-22.84 = -23.20 + 0.36)

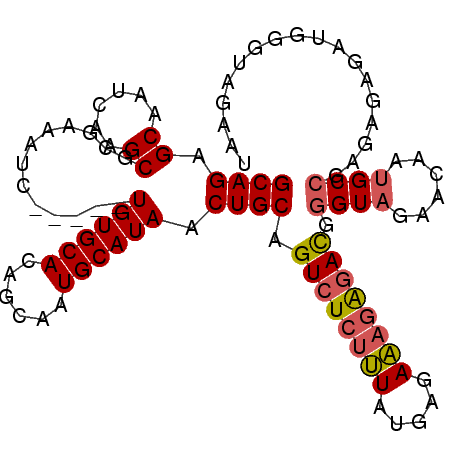
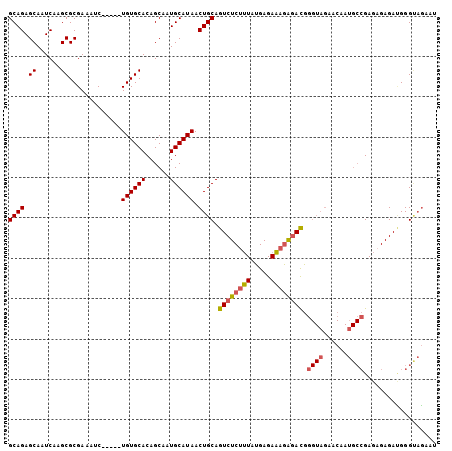
| Location | 6,458,469 – 6,458,571 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -27.53 |
| Consensus MFE | -23.62 |
| Energy contribution | -23.27 |
| Covariance contribution | -0.36 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.10 |
| Structure conservation index | 0.86 |
| SVM decision value | 1.13 |
| SVM RNA-class probability | 0.920546 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
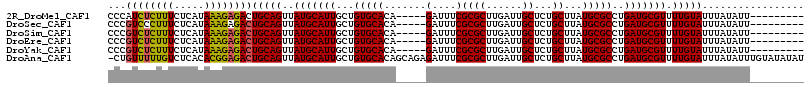
>2R_DroMel_CAF1 6458469 102 + 20766785 CCCAUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCAUUGCUGUGCACA-----GAUUUCGCGCUUGAUUGCUCUGCUUAUGCGCCUGAUGCGUUUUGUAUUUAUAUU--------- ....(((((((.....))))))).(((((..(((((((...(((((((-----((....(((......)))))))....)))))..))))))).)))))........--------- ( -24.30) >DroSec_CAF1 34102 102 + 1 CCCGUCCCUUUCUCAUAAAGAGACUGCAGUUAUGCAUUGCUGUGCACA-----GAUUUCGCGCUUGAUUGCUCUGCUUAUGCGCCUGAUGCGUUUUGUAUUUAUAUU--------- ...(((.((((.....)))).)))(((((..(((((((...(((((((-----((....(((......)))))))....)))))..))))))).)))))........--------- ( -22.90) >DroSim_CAF1 25465 102 + 1 CCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCAUUGCUGUGCACA-----GAUUUCGCGCUUGAUUGCUCUGCUUAUGCGCCUGAUGCGUUUUGUAUUUAUAUU--------- ...((((((((.....))))))))(((((..(((((((...(((((((-----((....(((......)))))))....)))))..))))))).)))))........--------- ( -27.70) >DroEre_CAF1 32422 102 + 1 CCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCAUUGCUGUGCACA-----GAUUUCGCGCUUGAUUGCUCUGCUUAUGCGCCUGAUGCGUUUUGUAUUUAUAUU--------- ...((((((((.....))))))))(((((..(((((((...(((((((-----((....(((......)))))))....)))))..))))))).)))))........--------- ( -27.70) >DroYak_CAF1 36065 102 + 1 CCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCAUUGCUGUGCACA-----GAUUUCGCGCUUGAUUGCUCUGCUUAUGCGCCUGAUGCGUUUUGUAUUUAUAUU--------- ...((((((((.....))))))))(((((..(((((((...(((((((-----((....(((......)))))))....)))))..))))))).)))))........--------- ( -27.70) >DroAna_CAF1 35595 115 + 1 -CUGUUUUUGUCUCACACGGAGACUGCAGUUAUGCAUUGCUGUGCACAGCAGAGAUUUCGCGCUUGAUUGCUCUGCUUAUGCGCCUGAUGCGUUUUGUAUUUAUAUUUGUAUAUAU -........(((((.....)))))(((((..(((((((...(((((.(((((((...(((....)))...)))))))..)))))..))))))).)))))..(((((....))))). ( -34.90) >consensus CCCGUCUCUUUCUCAUAAAGAGACUGCAGUUAUGCAUUGCUGUGCACA_____GAUUUCGCGCUUGAUUGCUCUGCUUAUGCGCCUGAUGCGUUUUGUAUUUAUAUU_________ ...((((((((.....))))))))(((((..(((((((...(((((.......(....)((((......))...))...)))))..))))))).)))))................. (-23.62 = -23.27 + -0.36)
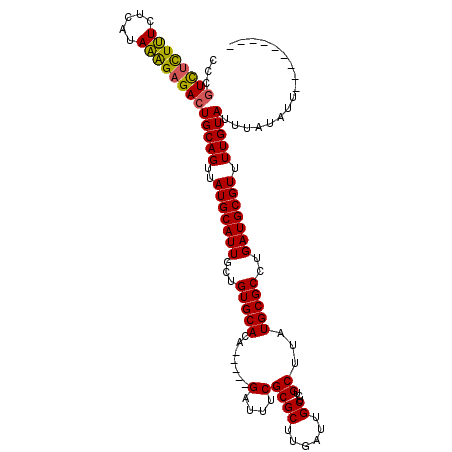
| Location | 6,458,469 – 6,458,571 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 92.50 |
| Mean single sequence MFE | -25.27 |
| Consensus MFE | -23.44 |
| Energy contribution | -23.38 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.93 |
| SVM decision value | 2.78 |
| SVM RNA-class probability | 0.996993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6458469 102 - 20766785 ---------AAUAUAAAUACAAAACGCAUCAGGCGCAUAAGCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGAUGGG ---------...............(((.....))).....((((.((......))........-----((((((......)))))).)))).((((((((.....))))))))... ( -23.20) >DroSec_CAF1 34102 102 - 1 ---------AAUAUAAAUACAAAACGCAUCAGGCGCAUAAGCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGGGACGGG ---------...............(((.....))).....((((.((......))........-----((((((......)))))).)))).((((((((.....))))))))... ( -24.50) >DroSim_CAF1 25465 102 - 1 ---------AAUAUAAAUACAAAACGCAUCAGGCGCAUAAGCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGG ---------...............(((.....))).....((((.((......))........-----((((((......)))))).)))).((((((((.....))))))))... ( -25.40) >DroEre_CAF1 32422 102 - 1 ---------AAUAUAAAUACAAAACGCAUCAGGCGCAUAAGCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGG ---------...............(((.....))).....((((.((......))........-----((((((......)))))).)))).((((((((.....))))))))... ( -25.40) >DroYak_CAF1 36065 102 - 1 ---------AAUAUAAAUACAAAACGCAUCAGGCGCAUAAGCAGAGCAAUCAAGCGCGAAAUC-----UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGG ---------...............(((.....))).....((((.((......))........-----((((((......)))))).)))).((((((((.....))))))))... ( -25.40) >DroAna_CAF1 35595 115 - 1 AUAUAUACAAAUAUAAAUACAAAACGCAUCAGGCGCAUAAGCAGAGCAAUCAAGCGCGAAAUCUCUGCUGUGCACAGCAAUGCAUAACUGCAGUCUCCGUGUGAGACAAAAACAG- .(((((....))))).........(((((.(((((((((.((((((...((......))...))))))))))).......((((....))))))))..)))))............- ( -27.70) >consensus _________AAUAUAAAUACAAAACGCAUCAGGCGCAUAAGCAGAGCAAUCAAGCGCGAAAUC_____UGUGCACAGCAAUGCAUAACUGCAGUCUCUUUAUGAGAAAGAGACGGG ........................(((.....))).....((((.((......)).............((((((......)))))).)))).((((((((.....))))))))... (-23.44 = -23.38 + -0.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:22 2006