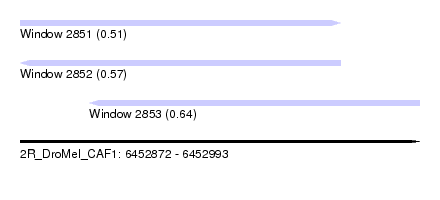
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,452,872 – 6,452,993 |
| Length | 121 |
| Max. P | 0.644473 |

| Location | 6,452,872 – 6,452,969 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -21.34 |
| Energy contribution | -21.45 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.514599 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6452872 97 + 20766785 ---UUGUGAUGGAUUAACCGGCGCUUAAGCUGCAGUCGAGCGAUAAGUGCCGCUCAUGGCAGGCCAUUUUGUGUGAUGCCGUUCCCUUUCUGAAUCACCU ---..((((((((.....(((((((((.(((.......)))..)))))))))...((((((...(((.....))).))))))......)))..))))).. ( -29.40) >DroVir_CAF1 19347 85 + 1 GUCGCGUGAUGGAUUAAACGGCUCUUAAGCUGCAGCUGAGCGAUAAGCGGCGCUCAUGGCCUGCUAUUUU-UGAGCUGCCACAGCU-------------- ...(((((...........((((....))))((((((.((.((((.((((.((.....)))))))))).)-).))))))))).)).-------------- ( -28.50) >DroPse_CAF1 30698 85 + 1 ---UUGUGAUGGAUUAACCGGCGCUUAAGCUGCAGUCGAGCGAUAAGUGCCGCUCAUGGCAGGCCAUUUUGUGUGCUGCCGCUCACUC------------ ---..(((((((......(((((((((.(((.......)))..))))))))).....((((.((......)).)))).))).))))..------------ ( -29.70) >DroMoj_CAF1 24783 87 + 1 GUCGUGUGAUGGAUUAAACGGCUCUUAAGCUGCAGCCGAGCGAUAAGCGACGCUCAUGGCAAGCUAUUUU-UGAGCUGCCAGUGGCAU------------ (((((.(((....))).)))))......((..(....(((((........))))).(((((.(((.....-..)))))))))..))..------------ ( -29.90) >DroAna_CAF1 25142 85 + 1 ---UUGUGAUGGAUUAACCGGCGCUUAAGCUGCAGUCGAGCGAUAAGUGCCGCUCAUGGCAGGCCAUUUUGUGUGCUGCCGCUCACUC------------ ---..(((((((......(((((((((.(((.......)))..))))))))).....((((.((......)).)))).))).))))..------------ ( -29.70) >DroPer_CAF1 29216 85 + 1 ---UUGUGAUGGAUUAACCGGCGCUUAAGCUGCAGUCGAGCGAUAAGUGCCGCUCAUGGCAGGCCAUUUUGUGUGCUGCCGCUCACUC------------ ---..(((((((......(((((((((.(((.......)))..))))))))).....((((.((......)).)))).))).))))..------------ ( -29.70) >consensus ___UUGUGAUGGAUUAACCGGCGCUUAAGCUGCAGUCGAGCGAUAAGUGCCGCUCAUGGCAGGCCAUUUUGUGUGCUGCCGCUCACUC____________ .....(((((((......((((......))))((((.(((((........)))))((((....)))).......))))))).)))).............. (-21.34 = -21.45 + 0.11)

| Location | 6,452,872 – 6,452,969 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 81.48 |
| Mean single sequence MFE | -26.90 |
| Consensus MFE | -17.80 |
| Energy contribution | -17.83 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.66 |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.568910 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6452872 97 - 20766785 AGGUGAUUCAGAAAGGGAACGGCAUCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAAGCGCCGGUUAAUCCAUCACAA--- ..(((((........(((..((((...((.....))...)))).(((.((((.((((..(((.......))).)))).)))).))).))))))))..--- ( -26.70) >DroVir_CAF1 19347 85 - 1 --------------AGCUGUGGCAGCUCA-AAAAUAGCAGGCCAUGAGCGCCGCUUAUCGCUCAGCUGCAGCUUAAGAGCCGUUUAAUCCAUCACGCGAC --------------(((((..((.(((..-.....)))......((((((........))))))))..)))))........................... ( -23.80) >DroPse_CAF1 30698 85 - 1 ------------GAGUGAGCGGCAGCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAAGCGCCGGUUAAUCCAUCACAA--- ------------..((((..(((((..((.....))..))))).(((.((((.((((..(((.......))).)))).)))).))).....))))..--- ( -28.80) >DroMoj_CAF1 24783 87 - 1 ------------AUGCCACUGGCAGCUCA-AAAAUAGCUUGCCAUGAGCGUCGCUUAUCGCUCGGCUGCAGCUUAAGAGCCGUUUAAUCCAUCACACGAC ------------..(((..((((((((..-.....))).))))).(((((........)))))))).((.(((....))).))................. ( -24.50) >DroAna_CAF1 25142 85 - 1 ------------GAGUGAGCGGCAGCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAAGCGCCGGUUAAUCCAUCACAA--- ------------..((((..(((((..((.....))..))))).(((.((((.((((..(((.......))).)))).)))).))).....))))..--- ( -28.80) >DroPer_CAF1 29216 85 - 1 ------------GAGUGAGCGGCAGCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAAGCGCCGGUUAAUCCAUCACAA--- ------------..((((..(((((..((.....))..))))).(((.((((.((((..(((.......))).)))).)))).))).....))))..--- ( -28.80) >consensus ____________GAGUGAGCGGCAGCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAAGCGCCGGUUAAUCCAUCACAA___ ...................((((.((..........))((((..((((((........))))))...)))).......)))).................. (-17.80 = -17.83 + 0.03)


| Location | 6,452,893 – 6,452,993 |
|---|---|
| Length | 100 |
| Sequences | 5 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 88.30 |
| Mean single sequence MFE | -36.08 |
| Consensus MFE | -28.42 |
| Energy contribution | -28.58 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.56 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.644473 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6452893 100 - 20766785 GCGGCAUGCGUGCCAUUUCGGGGCAGGUGAUUCAGAAAGGGAACGGCAUCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAA ((((((....))))....((((((((((.(((..(...(....)(......).)..))).))))))).(((((((......))))))).)))..)).... ( -32.80) >DroSec_CAF1 28625 100 - 1 GCGGCAGGCGUGCCAUUUCGGGGCAGGUGAUUCGGAAAGGGAACGGCAUCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAA (((((((((.((((.......)))).((((((((.........))).)))))........))))))).(((((((......)))))))...))....... ( -39.10) >DroEre_CAF1 27137 100 - 1 GCGGCAGGCGUGCCAUUUCGGGGCAGGUGAUUCGGAAAGGGAACGGCAUCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAA (((((((((.((((.......)))).((((((((.........))).)))))........))))))).(((((((......)))))))...))....... ( -39.10) >DroYak_CAF1 30343 100 - 1 GCGGCAGGCGUGCCAUUUCGGGGCAGGUGAUUCGGAAAGGGAGCGGCAUCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAA (((((((((.((((.......)))).((((((((.........))).)))))........))))))).(((((((......)))))))...))....... ( -38.10) >DroAna_CAF1 25163 82 - 1 GUUGCAGGUGUGGCAUCU------------------GAGUGAGCGGCAGCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAA ((((((((((((((.((.------------------....))...))..)))))...((((....))))((((((......))))))..))))))).... ( -31.30) >consensus GCGGCAGGCGUGCCAUUUCGGGGCAGGUGAUUCGGAAAGGGAACGGCAUCACACAAAAUGGCCUGCCAUGAGCGGCACUUAUCGCUCGACUGCAGCUUAA (((((((((((.((.(((((((........))))))).))..))(......)........))))))).(((((((......)))))))...))....... (-28.42 = -28.58 + 0.16)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:18 2006