
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,447,446 – 6,447,577 |
| Length | 131 |
| Max. P | 0.749487 |

| Location | 6,447,446 – 6,447,538 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 76.42 |
| Mean single sequence MFE | -27.59 |
| Consensus MFE | -13.61 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.49 |
| SVM decision value | 0.47 |
| SVM RNA-class probability | 0.749487 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
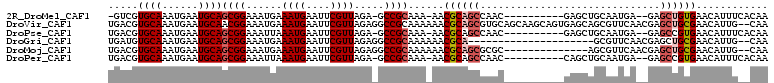
>2R_DroMel_CAF1 6447446 92 - 20766785 UAGA-GCCGCAAA-AACGCAGCCAAC----------GAGCUGCAAUGA--GAGCUGUGAACAUUUCACAAACGGUUUGCAAUUAAUGAGUGCGGACGUUCC----AAAAA ....-((((((..-...(((((....----------..)))))..((.--.((((((.............))))))..)).........))))).).....----..... ( -23.62) >DroVir_CAF1 14776 107 - 1 UAGAGGCCGCAAAAAACGCAGCGUGCAGCAAGCAGUGAGCAGCGUUCAACGAGCUGCGAACAUUG--CAAACGGUUUGCAAUUAAUGAGUGCCUGCCGCUAUGU-AAUAA ...............(((.((((.((((((..((((..(((((.........)))))..))((((--(((.....)))))))...))..)).)))))))).)))-..... ( -33.70) >DroPse_CAF1 24614 95 - 1 UAGA-GCCGCAAA-AACGCAGCCAAC----------GAGCUGCAAUGA--GAGCCGUGAACAUUUCACAAACGGUUCGCAAUUAAUGAGUGCGCGAGAGCAUCG-ACCAA ..((-...(((..-...(((((....----------..)))))..((.--(((((((.............))))))).)).........)))((....)).)).-..... ( -27.12) >DroMoj_CAF1 20150 93 - 1 UAGAGGCCGCAAAAAACGCAGCGCGC--------------AGCGUUCAACGAGCUGCGAACAUUG--CAAACGGUUCGCAAUUAAUGAGUGCCUGCUGCUAUGC-AAUAA ........(((......((((.((((--------------...((((...))))(((((((....--......)))))))........)))))))).....)))-..... ( -25.40) >DroAna_CAF1 20060 96 - 1 UAGA-GCCGCAAA-AACGCAGCCAAC----------GAGCUGCAAUGA--GAGCUGUGAACAUUUCUCAAACGGUUUGCAAUUAAUGAGUGCGAACGGCCCCCCAAAAAA ....-(((.....-...(((((....----------..)))))..(((--(((.((....)).))))))....(((((((.........))))))))))........... ( -28.60) >DroPer_CAF1 23118 95 - 1 UAGA-GCCGCAAA-AACGCAGCCAAC----------CAGCUGCAAUGA--GAGCCGUGAACAUUUCACAAACGGUUCGCAAUUAAUGAGUGCGCGAGAGCAUCG-ACCAA ..((-...(((..-...(((((....----------..)))))..((.--(((((((.............))))))).)).........)))((....)).)).-..... ( -27.12) >consensus UAGA_GCCGCAAA_AACGCAGCCAAC__________GAGCUGCAAUGA__GAGCUGUGAACAUUUCACAAACGGUUCGCAAUUAAUGAGUGCGCGCGGCCAUCC_AAAAA .......((((......(((((................))))).......(((((((.............)))))))............))))................. (-13.61 = -14.48 + 0.86)

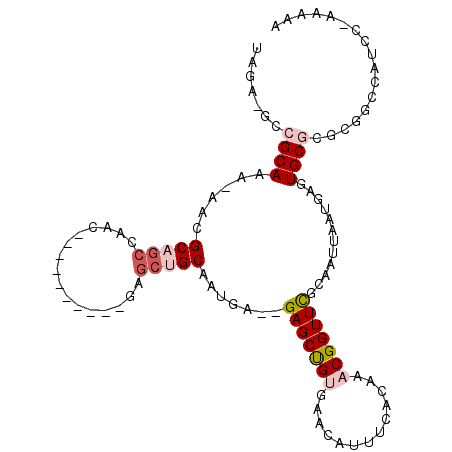
| Location | 6,447,482 – 6,447,577 |
|---|---|
| Length | 95 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.68 |
| Mean single sequence MFE | -26.87 |
| Consensus MFE | -15.23 |
| Energy contribution | -15.81 |
| Covariance contribution | 0.58 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.723449 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6447482 95 - 20766785 -GUCGUGCAAAUGAAUGCAGCGGAAAUGAAAUGAAUUCGUUAGA-GCCGCAAA-AACGCAGCCAAC----------GAGCUGCAAUGA--GAGCUGUGAACAUUUCACAA -...(((.(((((..((((((((.((((((.....))))))...-.)).....-...(((((....----------..))))).....--..))))))..)))))))).. ( -27.60) >DroVir_CAF1 14815 108 - 1 UGACGUGCAAAUGAAUGCAACGGAAAUGAAAUGAAUUCGUUAGAGGCCGCAAAAAACGCAGCGUGCAGCAAGCAGUGAGCAGCGUUCAACGAGCUGCGAACAUUG--CAA ...((((((......))).)))..((((((.....)))))).(..(((((..........))).))..)..((((((.(((((.........)))))...)))))--).. ( -30.30) >DroPse_CAF1 24653 96 - 1 UGACGUGCAAAUGAAUGCAGCGGAAAUUAAAUGAAUUCGUUAGA-GCCGCAAA-AACGCAGCCAAC----------GAGCUGCAAUGA--GAGCCGUGAACAUUUCACAA ....(((.(((((..(((.((((...((.((((....)))).))-.))))...-...(((((....----------..))))).....--.....)))..)))))))).. ( -25.60) >DroGri_CAF1 5287 87 - 1 UGAUGUGCAAAUGAAUGCAGCGGAAAUGAAAUGAAUUCGUUAGAGGCCGCAAAAAACGCA---------------------GCGUUCAACGAGCUGCGAACAUUG--CAA .....(((((.((......((((.((((((.....)))))).....))))......((((---------------------((.........))))))..)))))--)). ( -25.30) >DroMoj_CAF1 20189 94 - 1 UGACGUGCAAAUGAAUGCAGCGGAAAUGAAAUGAAUUCGUUAGAGGCCGCAAAAAACGCAGCGCGC--------------AGCGUUCAACGAGCUGCGAACAUUG--CAA .....((((......))))((((.((((((.....)))))).....)))).......((((..(((--------------(((.........))))))....)))--).. ( -26.80) >DroPer_CAF1 23157 96 - 1 UGACGUGCAAAUGAAUGCAGCGGAAAUUAAAUGAAUUCGUUAGA-GCCGCAAA-AACGCAGCCAAC----------CAGCUGCAAUGA--GAGCCGUGAACAUUUCACAA ....(((.(((((..(((.((((...((.((((....)))).))-.))))...-...(((((....----------..))))).....--.....)))..)))))))).. ( -25.60) >consensus UGACGUGCAAAUGAAUGCAGCGGAAAUGAAAUGAAUUCGUUAGA_GCCGCAAA_AACGCAGCCAAC__________GAGCUGCAAUCA__GAGCUGCGAACAUUG__CAA .....((((......))))((((......((((....)))).....))))......((((((..............................))))))............ (-15.23 = -15.81 + 0.58)
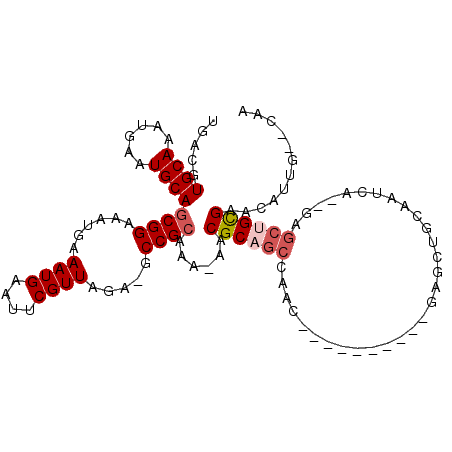

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:13 2006