
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,440,564 – 6,440,671 |
| Length | 107 |
| Max. P | 0.594592 |

| Location | 6,440,564 – 6,440,671 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -24.25 |
| Consensus MFE | -14.03 |
| Energy contribution | -13.78 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.594592 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
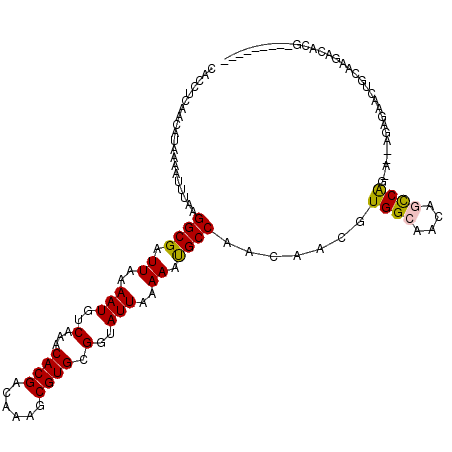
>2R_DroMel_CAF1 6440564 107 - 20766785 CACCUCAACAUAAAAUUUAAGGCGAUUAAAAUGUCAAACACGACAAAGCGUGCGGUAUUAAAAAUGCCAACAACGUGGCAACAGCCAGUAC-AGUGAGUUGCGGGGCG----------- ..((((..(...........)((((((....((((......))))....(((((((((.....))))).......((((....))))))))-....))))))))))..----------- ( -25.90) >DroVir_CAF1 8855 110 - 1 CACCUCAACAUAAAAUUUAAGGCGAUUAAAAUGUCAAACACGACAAAGCGUGCGGUAUUAAAAAUGCCAACAACGUGGCAACAAUCGC-----AAACACAGCCACACACACACAC---- ....................(((........((((......))))..(..((((((........(((((......)))))...)))))-----)..)...)))............---- ( -20.70) >DroYak_CAF1 14761 113 - 1 CACCUCAACAUAAAAUUUAAGGCGAUUAAAAUGUCAAACACGACAAAGCGUGCGGUAUUAAAAAUGCCAACAACGUGGCAACAGCCAGUAC-GGGGAACUGGAUGGGAGC-----GGGG ..((((.........................((((......))))..(((((((((((.....))))).......((((....))))))))-((....))........))-----)))) ( -26.50) >DroMoj_CAF1 13633 110 - 1 CACCUCAACAUAAAAUUUAAGGCGAUUAAAAUGUCAAACACGACAAAGCGUGCGGUAUUAAAAACGCCAACAACGUGGCAACAAUCAC-----ACUCACACAUACACACGCACAC---- (.(((.((........)).))).).......((((......))))..(((((.(((.......))((((......)))).........-----...........).)))))....---- ( -17.90) >DroAna_CAF1 13413 113 - 1 CACCUCAACAUAAAAUUUAAGGCGAUUAAAAUGUCAAACACGACAAAGCGUGCGGUAUUAAAAAUGCCAACAACGUGGCAACAGCCAACAGGAGAGAG-UGCAGGAUGAG-----AGGG ..((((.........................((((......))))...(.((((((((.....)))))..(....((((....))))....)......-.))).)....)-----))). ( -25.30) >DroPer_CAF1 13676 113 - 1 CACCUCAACAUAAAAUUUAAGGCGAUUAAAAUGUCAAACACGACAAAGCGUGCGGUAUUAAAAACGCCAACAGCGUGGCAACAGCCAGCAG-AGAGCACUGCCAAAGAGG-----GGGG ..((((....((.....)).((((.......((((......))))....(((((((.........)))....((.((((....))))))..-...))))))))...))))-----.... ( -29.20) >consensus CACCUCAACAUAAAAUUUAAGGCGAUUAAAAUGUCAAACACGACAAAGCGUGCGGUAUUAAAAAUGCCAACAACGUGGCAACAGCCAG_A__AGAGAACUGCAAGACACG_________ ....................((((.((..(((..(...((((......)))).)..)))..)).)))).......((((....))))................................ (-14.03 = -13.78 + -0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:10 2006