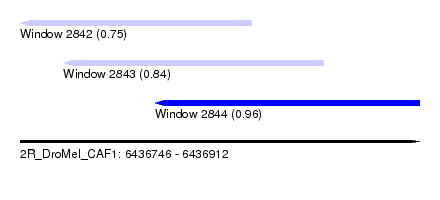
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,436,746 – 6,436,912 |
| Length | 166 |
| Max. P | 0.961289 |

| Location | 6,436,746 – 6,436,842 |
|---|---|
| Length | 96 |
| Sequences | 5 |
| Columns | 97 |
| Reading direction | reverse |
| Mean pairwise identity | 84.16 |
| Mean single sequence MFE | -22.64 |
| Consensus MFE | -12.94 |
| Energy contribution | -14.02 |
| Covariance contribution | 1.08 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.53 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.48 |
| SVM RNA-class probability | 0.753209 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6436746 96 - 20766785 AAAUAGCUGGAAACAUGAAGUGCAAUAUACAUAUAUGUAUGUAUGUACGGCA-GCAACAUCGAGUGCAACUUGCUCUCAUCGCCCAUCAAGGCAGCA .....((((....).....((((((((((((....))))))).)))))))).-((......(((.((.....)).)))...(((......))).)). ( -28.70) >DroSec_CAF1 12559 79 - 1 AAAUAGCUGGAAACAUGAAGUGCAAUAUA--------UAUGU---------A-GCAACAUCGAGUGCAACUUGCUCUCAUCGCCCAUCAAGGCAGCA .....((((....)..((..(((.(((..--------..)))---------.-)))...))(((.((.....)).)))...(((......)))))). ( -17.70) >DroSim_CAF1 11022 88 - 1 AAAUAGCUGGAAACAUGAAGUGCAAUAUA--------UAUGUAUGAACGGCA-GCAACAUCGAGUGCAACUUGCUCUCAUCGCCCAUCAAGGCAGCA .....((((....).....(((((.....--------..)))))....))).-((......(((.((.....)).)))...(((......))).)). ( -20.00) >DroEre_CAF1 11080 79 - 1 AAAUAGCUGGAAACAUGAAGUGCAAUAUA--------CA----------GCAGGCAACAUGCAGUGCAACUUGCACUCAUCGCCCAUCAAGGCAGCA .....((((....)((((.((((((..((--------(.----------((((....).))).)))....)))))))))).(((......)))))). ( -24.00) >DroYak_CAF1 11253 88 - 1 AAAUAGCUGGCAACAUGAAGUGCAAUAUA--------CACAGCUACAGAGCA-GCAACAUCCAGUGCAACUUGCACUCAUCGCCCAUCAAGGCAGCA .....((((....)((((.((((((....--------....(((....))).-(((........)))...)))))))))).(((......)))))). ( -22.80) >consensus AAAUAGCUGGAAACAUGAAGUGCAAUAUA________UAUGU_U__A__GCA_GCAACAUCGAGUGCAACUUGCUCUCAUCGCCCAUCAAGGCAGCA .....((((....)((((.(.(((((((((......)))))............(((........)))...)))).))))).(((......)))))). (-12.94 = -14.02 + 1.08)

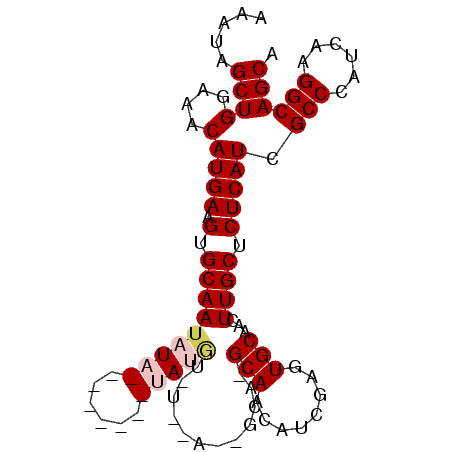
| Location | 6,436,764 – 6,436,872 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 82.85 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -10.72 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6436764 108 - 20766785 CAUCAAAAACUACG-AG--AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUACAUAUAUGUAUGUAUGUACGGCA-GCAACAUCGAGUGCAACUUGCUCUCA .............(-(.--........((((((....)))))).....((..((((((((((((....))))))).)))))..))-......))(((.((.....)).))). ( -25.80) >DroSec_CAF1 12577 90 - 1 CAUCAAAAACUACGA----AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA--------UAUGU---------A-GCAACAUCGAGUGCAACUUGCUCUCA ...............----........((((((....))))))......((..(((.(((..--------..)))---------.-)))...))(((.((.....)).))). ( -16.50) >DroSim_CAF1 11040 101 - 1 CAUCAAAAACUACAGAG--AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA--------UAUGUAUGAACGGCA-GCAACAUCGAGUGCAACUUGCUCUCA ..............(((--(.......((((((....)))))).......(((((((.....--------.((((.((.......-.))))))....))).))))..)))). ( -17.80) >DroEre_CAF1 11098 93 - 1 CAUCAAAAGCAACG-AGCAAAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA--------CA----------GCAGGCAACAUGCAGUGCAACUUGCACUCA ..(((...((....-.)).........((((((....)))))).....)))(((((((..((--------(.----------((((....).))).)))....))))))).. ( -22.90) >DroYak_CAF1 11271 100 - 1 CAUCAAAAACUACG-AG--AAAAAUAACCAGUUAAAUAGCUGGCAACAUGAAGUGCAAUAUA--------CACAGCUACAGAGCA-GCAACAUCCAGUGCAACUUGCACUCA ..............-..--........((((((....)))))).....(((.((((((....--------....(((....))).-(((........)))...))))))))) ( -19.80) >consensus CAUCAAAAACUACG_AG__AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA________UAUGU_U__A__GCA_GCAACAUCGAGUGCAACUUGCUCUCA ...........................((((((....))))))...................................................(((.((.....)).))). (-10.72 = -11.12 + 0.40)

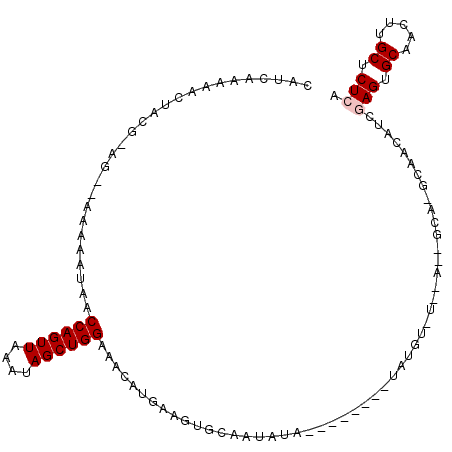
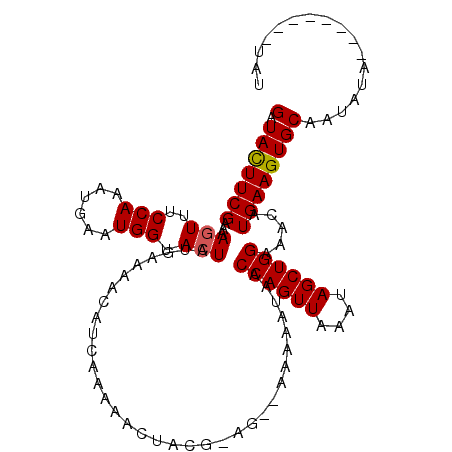
| Location | 6,436,802 – 6,436,912 |
|---|---|
| Length | 110 |
| Sequences | 5 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 90.07 |
| Mean single sequence MFE | -16.36 |
| Consensus MFE | -14.46 |
| Energy contribution | -14.42 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.52 |
| SVM RNA-class probability | 0.961289 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6436802 110 - 20766785 GAUACUUCGAAAAGUUUCCAAAUGAAUGGUACUAUGAAAACAUCAAAAACUACG-AG--AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUACAUAUAUGUAU (.(((((((...(((..(((......))).))).....................-..--........((((((....)))))).....))))))))...(((((....))))) ( -17.70) >DroSec_CAF1 12606 101 - 1 GAUACUUCGAAAAGUUUCCAAAUGAAUGGUACUAUGAAAACAUCAAAAACUACGA----AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA--------UAU (.(((((((...(((..(((......))).)))......................----........((((((....)))))).....))))))))......--------... ( -16.60) >DroSim_CAF1 11078 103 - 1 GAUACUUCGAAAAGUUUCCAAAUGAAUGGUACUAUGAAAACAUCAAAAACUACAGAG--AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA--------UAU (.(((((((...(((..(((......))).)))........................--........((((((....)))))).....))))))))......--------... ( -16.60) >DroEre_CAF1 11128 103 - 1 GGUAUUUCGAAAAGUUUCCAAAUGAAUGGCACUAUGAAAACAUCAAAAGCAACG-AGCAAAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA--------CA- .((((((((....(((((((..((..((((....(((.....)))...((....-.))............))))..))..))))))).))))))))......--------..- ( -18.80) >DroYak_CAF1 11309 101 - 1 GAUAUUUCGAA-ACUUUCCAAAUGAAUGGUACUAUGAAAACAUCAAAAACUACG-AG--AAAAAUAACCAGUUAAAUAGCUGGCAACAUGAAGUGCAAUAUA--------CAC ..(((((.((.-....)).)))))....(((((.(((.....))).........-..--........((((((....))))))........)))))......--------... ( -12.10) >consensus GAUACUUCGAAAAGUUUCCAAAUGAAUGGUACUAUGAAAACAUCAAAAACUACG_AG__AAAAAUAACCAGUUAAAUAGCUGGAAACAUGAAGUGCAAUAUA________UAU (.(((((((...(((..(((......))).)))..................................((((((....)))))).....))))))))................. (-14.46 = -14.42 + -0.04)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:10 2006