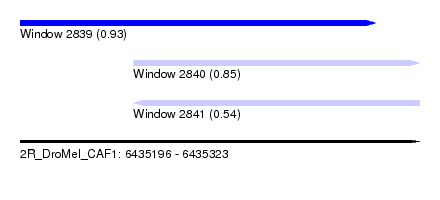
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,435,196 – 6,435,323 |
| Length | 127 |
| Max. P | 0.927417 |

| Location | 6,435,196 – 6,435,309 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 80.35 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -18.92 |
| Energy contribution | -19.95 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.22 |
| Mean z-score | -2.74 |
| Structure conservation index | 0.58 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.927417 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
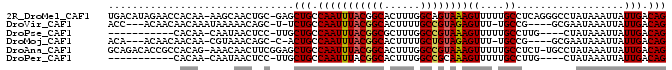
>2R_DroMel_CAF1 6435196 113 + 20766785 GUA---CGAACCGCAAGUGACUUUAGUGAAAUUUACAAGCUGUCAAUAAUUUAUAGGCCCUGAGGCAAAAACUUUACUGCCAAAGUGCCGUAAAUUGGCAGCUC-GCAGUUGCUU-UU ...---......((((.((......((((...)))).((((((((((...((((.(((.((..((((..........))))..)).))))))))))))))))).-.)).))))..-.. ( -32.10) >DroVir_CAF1 4609 108 + 1 UCA---CAAGCCGCAGCUGACUUUAGUGAAAUUUACAAGCUGUCAAUAAUUUAUUCGC----CGGCA-AAACUCUACGGCAAAAGUGCCGUAAAUUGGCAGA-A-GCUGUUUUUAUUU ...---......((((((.......((((...))))...((((((((.........(.----...).-......(((((((....))))))).)))))))).-)-)))))........ ( -26.30) >DroPse_CAF1 8799 112 + 1 GCGGGAGACACCGCAAGUGACUUUAGUGAAAUUUACAAGCUGUCAAUAAUUUAUAG----CAAGGCAAAAACUUUACGGCCAAAGCGCCGUAAAUUGGCAGCAA-GGAGUUAUUG-UU ((((......)))).(((((((((.((((...))))..(((((((((........(----(...)).......(((((((......))))))))))))))))..-))))))))).-.. ( -37.40) >DroMoj_CAF1 9011 107 + 1 ACA---GAAGCCACAGCUGACUUUAGUGAAAUUUACAAGCUGUCAAUAAUUUAUUCGC----CGGCA-AAACUCUACAGCAAAAGUGCCGUAAAUUGGCAGU-G-GCUGUUUACG-UU ...---..((((((.((((......((((...))))..((((.(((((...)))).).----)))).-........)))).....(((((.....)))))))-)-))).......-.. ( -27.60) >DroAna_CAF1 9527 113 + 1 GGG---ACUACCGCAAGUGACUUUAGUGAAAUUUACAAGCUGUCAAUAAUUUAUAGGCA-AGAGGCAAAAACUUUACGGCCAAAGUGCCGUAAAUUGGCAGCUCCGAAGUUGUUU-CU ((.---....))(.(((..(((((.((((...)))).((((((((((.........((.-....)).......(((((((......)))))))))))))))))..)))))..)))-). ( -32.40) >DroPer_CAF1 9042 112 + 1 GCGGGAGACACCGCAAGUGACUUUAGUGAAAUUUACAAGCUGUCAAUAAUUUAUAG----CAAGGCAAAAACUUUGCGGCCAAAGUGCCGUAAAUUGGCAGCAA-GGAGUUAUUG-UU ((((......)))).(((((((((.((((...))))..(((((((((........(----(((((......))))))(((......)))....)))))))))..-))))))))).-.. ( -39.10) >consensus GCA___GAAACCGCAAGUGACUUUAGUGAAAUUUACAAGCUGUCAAUAAUUUAUAGGC____AGGCAAAAACUUUACGGCCAAAGUGCCGUAAAUUGGCAGC_A_GCAGUUAUUG_UU ...............((((((....((((...))))..(((((((((.........((......))........((((((......)))))).)))))))))......)))))).... (-18.92 = -19.95 + 1.03)

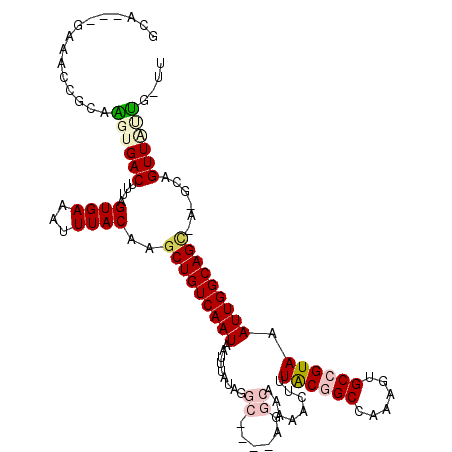
| Location | 6,435,232 – 6,435,323 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -21.85 |
| Consensus MFE | -13.45 |
| Energy contribution | -14.32 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.847532 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6435232 91 + 20766785 CUGUCAAUAAUUUAUAGGCCCUGAGGCAAAAACUUUACUGCCAAAGUGCCGUAAAUUGGCAGCUC-GCAGUUGCUU-UUGUGGUUCUAUGUCA ............(((((..((.((((((........(((((...(((((((.....))))).)).-))))))))))-)...))..)))))... ( -25.00) >DroVir_CAF1 4645 83 + 1 CUGUCAAUAAUUUAUUCGC----CGGCA-AAACUCUACGGCAAAAGUGCCGUAAAUUGGCAGA-A-GCUGUUUUUAUUUGUUGUUGU---GGU ...............((((----(((((-((.....(((((.....(((((.....)))))..-.-))))).....)))))))..))---)). ( -18.80) >DroPse_CAF1 8838 76 + 1 CUGUCAAUAAUUUAUAG----CAAGGCAAAAACUUUACGGCCAAAGCGCCGUAAAUUGGCAGCAA-GGAGUUAUUG-UUGUG----------- ....(((((((((...(----(...((...((.((((((((......)))))))))).)).))..-.)))))))))-.....----------- ( -20.80) >DroMoj_CAF1 9047 82 + 1 CUGUCAAUAAUUUAUUCGC----CGGCA-AAACUCUACAGCAAAAGUGCCGUAAAUUGGCAGU-G-GCUGUUUACG-UUGUUGUUGU---UGU ....((((((.........----(((((-(......(((((...(.(((((.....))))).)-.-))))).....-))))))))))---)). ( -21.10) >DroAna_CAF1 9563 91 + 1 CUGUCAAUAAUUUAUAGGCA-AGAGGCAAAAACUUUACGGCCAAAGUGCCGUAAAUUGGCAGCUCCGAAGUUGUUU-CUGUGGCGGUGUCUGC ((((((.((((((((.((((-...(((............)))....))))))))))))((((((....))))))..-...))))))....... ( -24.10) >DroPer_CAF1 9081 76 + 1 CUGUCAAUAAUUUAUAG----CAAGGCAAAAACUUUGCGGCCAAAGUGCCGUAAAUUGGCAGCAA-GGAGUUAUUG-UUGUG----------- .......((((((((.(----(((((......))))))(((......)))))))))))(((((((-.......)))-)))).----------- ( -21.30) >consensus CUGUCAAUAAUUUAUAGGC____AGGCAAAAACUUUACGGCCAAAGUGCCGUAAAUUGGCAGC_A_GCAGUUAUUG_UUGUGGUUGU___UG_ ((((((((.........((......))........((((((......)))))).))))))))............................... (-13.45 = -14.32 + 0.86)

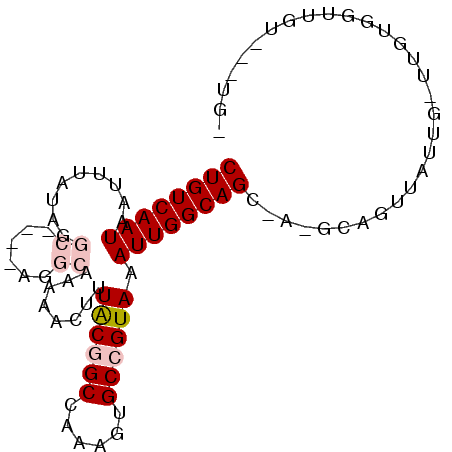
| Location | 6,435,232 – 6,435,323 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 74.15 |
| Mean single sequence MFE | -17.22 |
| Consensus MFE | -10.83 |
| Energy contribution | -10.83 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.18 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.544430 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
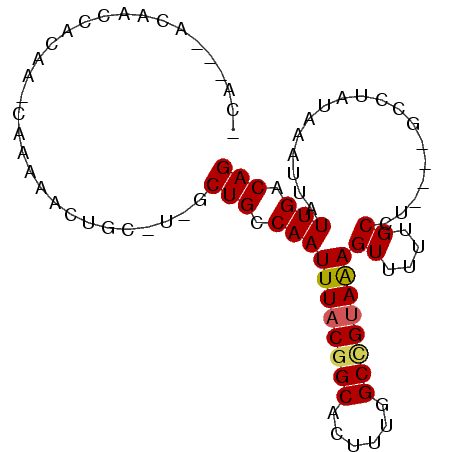
>2R_DroMel_CAF1 6435232 91 - 20766785 UGACAUAGAACCACAA-AAGCAACUGC-GAGCUGCCAAUUUACGGCACUUUGGCAGUAAAGUUUUUGCCUCAGGGCCUAUAAAUUAUUGACAG ....((((..((.(((-((((.(((((-(((.((((.......)))))))..)))))...))))))).....))..))))............. ( -22.30) >DroVir_CAF1 4645 83 - 1 ACC---ACAACAACAAAUAAAAACAGC-U-UCUGCCAAUUUACGGCACUUUUGCCGUAGAGUUU-UGCCG----GCGAAUAAAUUAUUGACAG ...---...................((-(-...((...(((((((((....)))))))))....-.)).)----))................. ( -15.70) >DroPse_CAF1 8838 76 - 1 -----------CACAA-CAAUAACUCC-UUGCUGCCAAUUUACGGCGCUUUGGCCGUAAAGUUUUUGCCUUG----CUAUAAAUUAUUGACAG -----------.....-((((((....-..((.((...((((((((......))))))))......))...)----)......)))))).... ( -17.10) >DroMoj_CAF1 9047 82 - 1 ACA---ACAACAACAA-CGUAAACAGC-C-ACUGCCAAUUUACGGCACUUUUGCUGUAGAGUUU-UGCCG----GCGAAUAAAUUAUUGACAG ...---..........-........((-(-...((...(((((((((....)))))))))....-.)).)----))................. ( -15.40) >DroAna_CAF1 9563 91 - 1 GCAGACACCGCCACAG-AAACAACUUCGGAGCUGCCAAUUUACGGCACUUUGGCCGUAAAGUUUUUGCCUCU-UGCCUAUAAAUUAUUGACAG (((((.((.((..(.(-((.....))))..))......((((((((......)))))))))).)))))....-.................... ( -18.50) >DroPer_CAF1 9081 76 - 1 -----------CACAA-CAAUAACUCC-UUGCUGCCAAUUUACGGCACUUUGGCCGCAAAGUUUUUGCCUUG----CUAUAAAUUAUUGACAG -----------.....-((((((..((-..(.((((.......)))).)..))..((((.((....)).)))----)......)))))).... ( -14.30) >consensus _CA___ACAACCACAA_CAAAAACUGC_U_GCUGCCAAUUUACGGCACUUUGGCCGUAAAGUUUUUGCCU____GCCUAUAAAUUAUUGACAG ...............................(((.(((((((((((......))))))))((....))..................))).))) (-10.83 = -10.83 + 0.00)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:07 2006