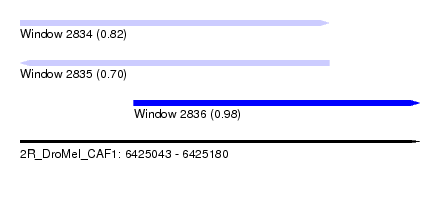
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,425,043 – 6,425,180 |
| Length | 137 |
| Max. P | 0.975687 |

| Location | 6,425,043 – 6,425,149 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -30.22 |
| Consensus MFE | -19.05 |
| Energy contribution | -19.33 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.824972 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
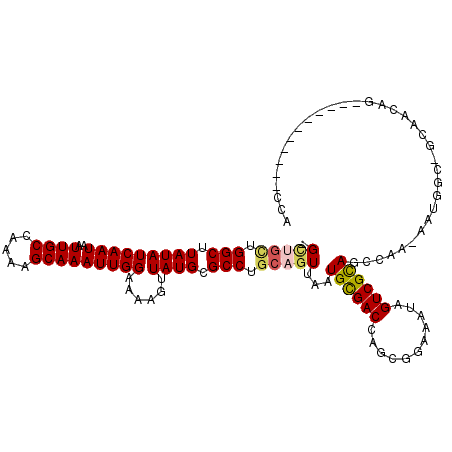
>2R_DroMel_CAF1 6425043 106 + 20766785 -GUUGCUGGCUUAUAUCAAUAAUUGCCAAAAGCAAAUUGGAAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCAGUCAA-AAUGGC-GCAACAA-----------CCA -((((.((((((...(((((..((((.....)))))))))..))))))((((((.(((.....)))(((..((((......)))).)))..-...)))-))).)))-----------).. ( -30.60) >DroVir_CAF1 46693 118 + 1 -GCCGCUGGCUUAUAUCAAUAAUUGCCAAAAGCAAAUUGGAAAAGUUAUGCGCCAGCAGUUAAUGUGACCAGCGAAAAUAGUCGCAACCCGUAACUGCAGCACCAGCAACAG-CAGCCGC -((.(((((((.......((((((.((((.......))))...))))))..((..(((((((.((((((...........)))))).....))))))).))...)))..)))-).))... ( -30.70) >DroPse_CAF1 5381 107 + 1 -GCUGUUGGCUUAUAUCAAUAAUUGCCAAAAGCAAAUUGGAAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCAGCCAAAAAUGGC-GCAACAG-----------CCG -(((((((((((...(((((..((((.....)))))))))..))))))((((((.........((((((...........)))))).........)))-)))))))-----------).. ( -33.17) >DroGri_CAF1 14665 86 + 1 UGGCAGUGGCUUAUAUCAAUAAUUGCCAAAAGCAAAUUGGAAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCA---------------------------------- .(((.(((((((...(((((..((((.....)))))))))..)))))))..)))((((.....))))....((((......)))).---------------------------------- ( -21.70) >DroMoj_CAF1 10659 119 + 1 -GCUGCCGGCUUAUAUCAAUAAUUGCCAAAAGCAAAUUGGAAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGUAGCCCAUAACAGCAGCAGCAGCAGCAGUCAGACUA -(((((.(((((...(((((..((((.....)))))))))..))))).(((..((((.((((.((((((...........)))))).....)))).))))..)))))))).......... ( -32.00) >DroPer_CAF1 12412 107 + 1 -GCUGUUGGCUUAUAUCAAUAAUUGCCAAAAGCAAAUUGGAAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCAGCCAAAAAUGGC-GCAACAG-----------CCG -(((((((((((...(((((..((((.....)))))))))..))))))((((((.........((((((...........)))))).........)))-)))))))-----------).. ( -33.17) >consensus _GCUGCUGGCUUAUAUCAAUAAUUGCCAAAAGCAAAUUGGAAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCAGCCAA_AAUGGC_GCAACAG___________CCA .(((((.(((.(((((((((..((((.....)))))))))......)))).))).)))))...((((((...........)))))).................................. (-19.05 = -19.33 + 0.28)


| Location | 6,425,043 – 6,425,149 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.62 |
| Mean single sequence MFE | -32.12 |
| Consensus MFE | -17.38 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.39 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.24 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696350 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
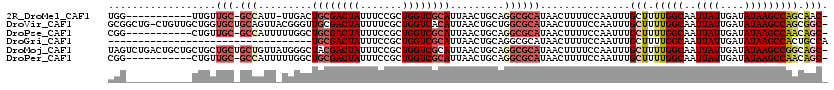
>2R_DroMel_CAF1 6425043 106 - 20766785 UGG-----------UUGUUGC-GCCAUU-UUGACUGCGACUAUUUCCGCUGGUCGCAUUAACUGCAGGCGCAUAACUUUUCCAAUUUGCUUUUGGCAAUUAUUGAUAUAAGCCAGCAAC- .((-----------((((.((-(((...-..(((((((........))).))))(((.....))).)))))))))))........(((((...(((..((((....)))))))))))).- ( -32.30) >DroVir_CAF1 46693 118 - 1 GCGGCUG-CUGUUGCUGGUGCUGCAGUUACGGGUUGCGACUAUUUUCGCUGGUCACAUUAACUGCUGGCGCAUAACUUUUCCAAUUUGCUUUUGGCAAUUAUUGAUAUAAGCCAGCGGC- ...((((-(((..(((.((((((((((((..(..(((((......)))).)..)....))))))).)))))..........((((((((.....))))..)))).....))))))))))- ( -38.20) >DroPse_CAF1 5381 107 - 1 CGG-----------CUGUUGC-GCCAUUUUUGGCUGCGACUAUUUCCGCUGGUCGCAUUAACUGCAGGCGCAUAACUUUUCCAAUUUGCUUUUGGCAAUUAUUGAUAUAAGCCAACAGC- ..(-----------(((((((-((((....))))((((((((.......))))))))......)).(((................((((.....))))((((....)))))))))))))- ( -32.70) >DroGri_CAF1 14665 86 - 1 ----------------------------------UGCGACUAUUUCCGCUGGUCGCAUUAACUGCAGGCGCAUAACUUUUCCAAUUUGCUUUUGGCAAUUAUUGAUAUAAGCCACUGCCA ----------------------------------((((((((.......)))))))).........((((((..............)))...((((..((((....))))))))..))). ( -21.24) >DroMoj_CAF1 10659 119 - 1 UAGUCUGACUGCUGCUGCUGCUGCUGUUAUGGGCUACGACUAUUUCCGCUGGUCGCAUUAACUGCAGGCGCAUAACUUUUCCAAUUUGCUUUUGGCAAUUAUUGAUAUAAGCCGGCAGC- .............(((((((..(((((((((.(((..(((((.......)))))(((.....))).))).)))))).....((((((((.....))))..)))).....))))))))))- ( -35.60) >DroPer_CAF1 12412 107 - 1 CGG-----------CUGUUGC-GCCAUUUUUGGCUGCGACUAUUUCCGCUGGUCGCAUUAACUGCAGGCGCAUAACUUUUCCAAUUUGCUUUUGGCAAUUAUUGAUAUAAGCCAACAGC- ..(-----------(((((((-((((....))))((((((((.......))))))))......)).(((................((((.....))))((((....)))))))))))))- ( -32.70) >consensus CGG___________CUGUUGC_GCCAUU_UUGGCUGCGACUAUUUCCGCUGGUCGCAUUAACUGCAGGCGCAUAACUUUUCCAAUUUGCUUUUGGCAAUUAUUGAUAUAAGCCAGCAGC_ ..................(((.(((.........((((((((.......)))))))).........))))))...............(((.(((((..((((....))))))))).))). (-17.38 = -18.77 + 1.39)

| Location | 6,425,082 – 6,425,180 |
|---|---|
| Length | 98 |
| Sequences | 4 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 72.10 |
| Mean single sequence MFE | -26.25 |
| Consensus MFE | -15.83 |
| Energy contribution | -15.52 |
| Covariance contribution | -0.31 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.02 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.75 |
| SVM RNA-class probability | 0.975687 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
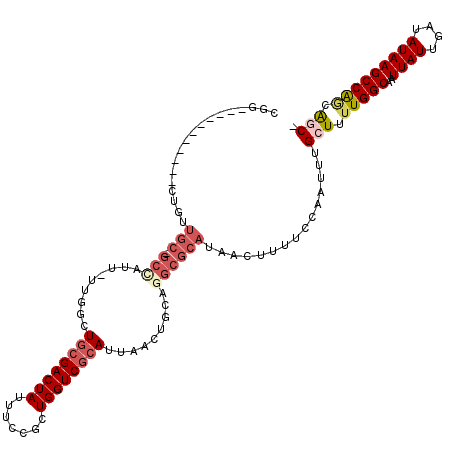
>2R_DroMel_CAF1 6425082 98 + 20766785 AAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCAGUCAA-AAUGGC-GCAACAA-----------CCAAACGGAAGUGCCCCAAACGCCCACA--CCCACA ....(((.((((((.(((.....)))(((..((((......)))).)))..-...)))-)))....-----------....((....))......))).......--...... ( -24.40) >DroVir_CAF1 46732 111 + 1 AAAAGUUAUGCGCCAGCAGUUAAUGUGACCAGCGAAAAUAGUCGCAACCCGUAACUGCAGCACCAGCAACAG-CAGCCGCAGCGGAAGUAUUAUAA-UACAUACAUCCACACA ........((((.(.(((((((.((((((...........)))))).....))))))).((....)).....-..).))))(.(((.((((.....-...)))).))).)... ( -27.20) >DroSec_CAF1 5519 90 + 1 AAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCAGUCAA-AAUGGC-GCAACAA-----------CCAAACGGAAGUGCCCCAAACACCCA---------- ....(((.((((((.(((.....)))(((..((((......)))).)))..-...)))-)))....-----------....((....))......))).....---------- ( -24.40) >DroMoj_CAF1 10698 110 + 1 AAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGUAGCCCAUAACAGCAGCAGCAGCAGCAGUCAGACUAAGCGGAAGCACCGCAA-UGCACACA--CAUACA ....((.(((.(..((((((....)).....((((...(((((((.((........)).)).((....)).....))))).((....)).))))..-))))..).--))))). ( -29.00) >consensus AAAAGUUAUGCGCCUGCAGUUAAUGCGACCAGCGGAAAUAGUCGCAGCCAA_AACGGC_GCAACAA___________CCAAACGGAAGUACCCCAA_CACACACA__CACACA ........((((((.........((((((...........)))))).........))).)))...................((....))........................ (-15.83 = -15.52 + -0.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:11:02 2006