
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,419,113 – 6,419,393 |
| Length | 280 |
| Max. P | 0.999980 |

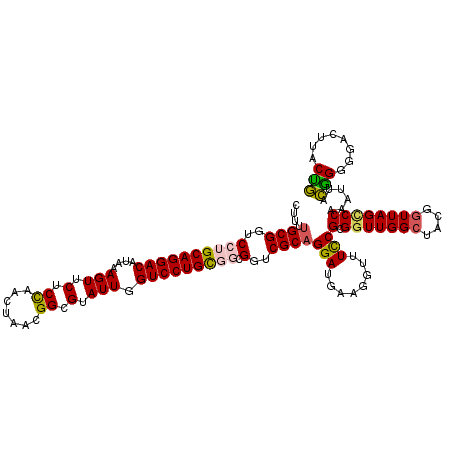
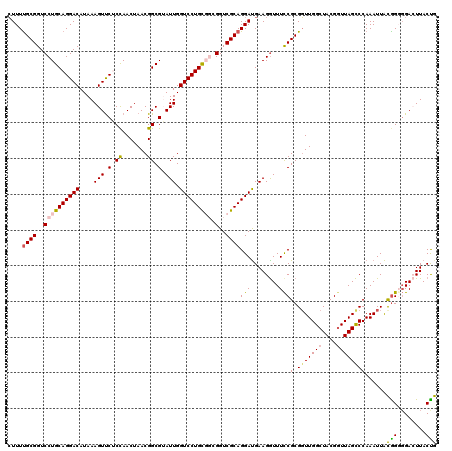
| Location | 6,419,113 – 6,419,233 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.97 |
| Mean single sequence MFE | -45.28 |
| Consensus MFE | -35.24 |
| Energy contribution | -35.00 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.78 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.915637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419113 120 - 20766785 CUUUUGCGGUCCUGCAGGACAUAAAGUUCUCCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUUCGCGGUUGGCUACGGUUAGCCCAAAUUACGGGCGACUUACUG .(((((((..((((((((((....(((.(.((.......)).).))).))))))))).)..)))))))....(((..(((((((((((....)))))))(........)))))...))). ( -45.80) >DroSec_CAF1 16670 120 - 1 CUUUUGCGGUCCUGCAGGACAUAAAGUUCACCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUCCGCGGUUGGCUACGUUUAGCCCAAAUUACGGGGGACUUACUG .(((((((..((((((((((....(((.(.((.......)).).))).))))))))).)..)))))))..(((.(..(((..((((((((....)))))..)))..)))..).))).... ( -47.80) >DroSim_CAF1 17357 120 - 1 CUUUUGCGGUCCUGCAGGACAUAAAGUUCACCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUCCGCGGUUGGCUACGGUUAGCCCAAAUUACAGGGGACUUACUG .(((((((..((((((((((....(((.(.((.......)).).))).))))))))).)..)))))))...((((..((..(((((((....))))))).........))..)))).... ( -48.30) >DroEre_CAF1 13847 117 - 1 CUUUUGCGGUCCUGCAGGACAUAAAGUCCUCUAACUAACGGCGGAUUGGUCCUGU---GGUCGCAGGAUGAAGAUUUCCGCGGUUGGCUACGGUUAGUCCAAAUUAUGGUGGAAUUACCA ....((((....))))((((.....)))).....(((((.(((((((.(((((((---....))))))).))....))))).)))))....(((...((((........))))...))). ( -42.60) >DroYak_CAF1 14317 117 - 1 GUUUCGCGGUCCUGCAGGACAUAAAGUUCUCUAACUAACGGCGGAUUGGUCCUGU---GGUCGCAGGACGAAGGUUUCCGCGCUUGGCUACGGUUAGCCCAAAUUGUGGUGGAAUUACCA (((..((((.((.(((((((....(((((.((.......)).))))).)))))))---))))))..)))...(((((((((((..(((((....)))))......)).))))))..))). ( -41.90) >consensus CUUUUGCGGUCCUGCAGGACAUAAAGUUCUCCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUCCGCGGUUGGCUACGGUUAGCCCAAAUUACGGGGGACUUACUG ....((((..((((((((((....(((.(.((.......)).).))).))))))))).)..))))(((........)))(.(((((((....))))))))......(((........))) (-35.24 = -35.00 + -0.24)

| Location | 6,419,153 – 6,419,273 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.49 |
| Mean single sequence MFE | -38.11 |
| Consensus MFE | -32.83 |
| Energy contribution | -33.55 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.97 |
| SVM RNA-class probability | 0.891207 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419153 120 - 20766785 CAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCUCCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUUCG ..(((((.................))))).......(((..(((((((..((((((((((....(((.(.((.......)).).))).))))))))).)..)))))))....)))..... ( -39.13) >DroSec_CAF1 16710 120 - 1 CAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCACCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUCCG ..(((((.................))))).......(((..(((((((..((((((((((....(((.(.((.......)).).))).))))))))).)..)))))))....)))..... ( -39.53) >DroSim_CAF1 17397 120 - 1 CAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCACCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUCCG ..(((((.................))))).......(((..(((((((..((((((((((....(((.(.((.......)).).))).))))))))).)..)))))))....)))..... ( -39.53) >DroEre_CAF1 13887 117 - 1 CAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUCCUCUAACUAACGGCGGAUUGGUCCUGU---GGUCGCAGGAUGAAGAUUUCCG ..(((((.................)))))...............(((((.((.(((((((....(((((.((.......)).))))).)))))))---)))))))(((........))). ( -35.93) >DroYak_CAF1 14357 117 - 1 CAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCGUUUCGCGGUCCUGCAGGACAUAAAGUUCUCUAACUAACGGCGGAUUGGUCCUGU---GGUCGCAGGACGAAGGUUUCCG ..(((((.................))))).......(((((((..((((.((.(((((((....(((((.((.......)).))))).)))))))---))))))..))))..)))..... ( -36.43) >consensus CAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCUCCAACUAACGGCGUAUUGGUCCUGCGGCGGUCGCAGGAUGAAGGUUUCCG ..(((((.................))))).......(((..(((((((..((((((((((....(((.(.((.......)).).))).))))))))).)..)))))))....)))..... (-32.83 = -33.55 + 0.72)

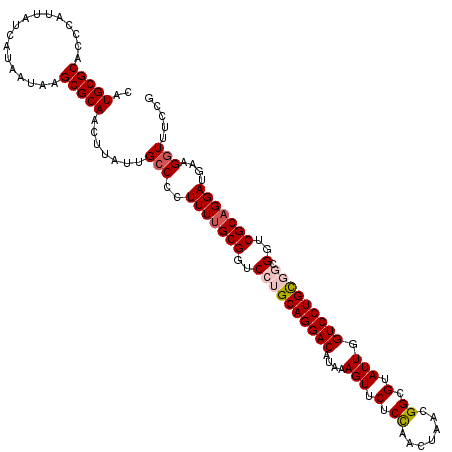
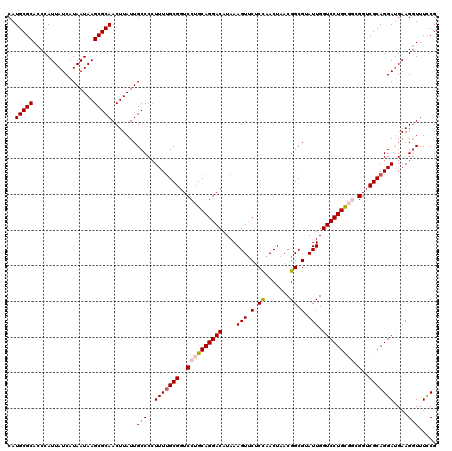
| Location | 6,419,193 – 6,419,313 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -37.86 |
| Consensus MFE | -37.78 |
| Energy contribution | -37.62 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.15 |
| Structure conservation index | 1.00 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.694002 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419193 120 + 20766785 CGUUAGUUGGAGAACUUUAUGUCCUGCAGGACCGCAAAAGGGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGC .............(((....(((((((.((.((.(((((..(((........((((((((((...))))))))))((....)).))).)))))...)).))...)))))))...)))... ( -37.90) >DroSec_CAF1 16750 120 + 1 CGUUAGUUGGUGAACUUUAUGUCCUGCAGGACCGCAAAAGGGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGC .............(((....(((((((.((.((.(((((..(((........((((((((((...))))))))))((....)).))).)))))...)).))...)))))))...)))... ( -37.90) >DroSim_CAF1 17437 120 + 1 CGUUAGUUGGUGAACUUUAUGUCCUGCAGGACCGCAAAAGGGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGC .............(((....(((((((.((.((.(((((..(((........((((((((((...))))))))))((....)).))).)))))...)).))...)))))))...)))... ( -37.90) >DroEre_CAF1 13924 120 + 1 CGUUAGUUAGAGGACUUUAUGUCCUGCAGGACCGCAAAAGGGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUAGC .............(((....(((((((.((.((.(((((..(((........((((((((((...))))))))))((....)).))).)))))...)).))...)))))))...)))... ( -38.00) >DroYak_CAF1 14394 120 + 1 CGUUAGUUAGAGAACUUUAUGUCCUGCAGGACCGCGAAACGGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGC .............(((....(((((((....(((.....))).......((.((((((((((...))))))))))))...(((((((..........))))))))))))))...)))... ( -37.60) >consensus CGUUAGUUGGAGAACUUUAUGUCCUGCAGGACCGCAAAAGGGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGC .............(((....(((((((.((.((.(((((..(((........((((((((((...))))))))))((....)).))).)))))...)).))...)))))))...)))... (-37.78 = -37.62 + -0.16)

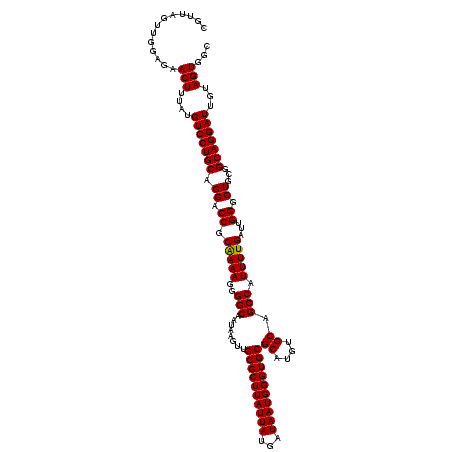

| Location | 6,419,193 – 6,419,313 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.67 |
| Mean single sequence MFE | -28.47 |
| Consensus MFE | -27.75 |
| Energy contribution | -27.75 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.97 |
| SVM decision value | 3.83 |
| SVM RNA-class probability | 0.999645 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419193 120 - 20766785 GCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCUCCAACUAACG ..........((((((.((((((..........))))))...(((((.................))))).......(((.(....).)))...)))))).....((((....)))).... ( -28.03) >DroSec_CAF1 16750 120 - 1 GCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCACCAACUAACG ..........((((((.((((((..........))))))...(((((.................))))).......(((.(....).)))...)))))).....((((....)))).... ( -28.03) >DroSim_CAF1 17437 120 - 1 GCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCACCAACUAACG ..........((((((.((((((..........))))))...(((((.................))))).......(((.(....).)))...)))))).....((((....)))).... ( -28.03) >DroEre_CAF1 13924 120 - 1 GCUACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUCCUCUAACUAACG ((.((..(((....((.((((((..........))))))...(((((.................))))).......)).....)))..))...))(((((.....))))).......... ( -28.63) >DroYak_CAF1 14394 120 - 1 GCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCGUUUCGCGGUCCUGCAGGACAUAAAGUUCUCUAACUAACG ..........((((((.((((((..........))))))...(((((.................))))).........(((.....)))....)))))).....((((....)))).... ( -29.63) >consensus GCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCCCUUUUGCGGUCCUGCAGGACAUAAAGUUCUCCAACUAACG ..........((((((.((((((..........))))))...(((((.................)))))....(((((.......)))))...))))))..................... (-27.75 = -27.75 + 0.00)


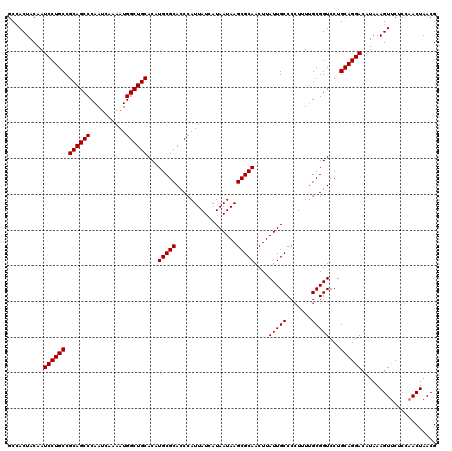
| Location | 6,419,233 – 6,419,353 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -42.26 |
| Consensus MFE | -39.66 |
| Energy contribution | -40.50 |
| Covariance contribution | 0.84 |
| Combinations/Pair | 1.02 |
| Mean z-score | -3.17 |
| Structure conservation index | 0.94 |
| SVM decision value | 4.45 |
| SVM RNA-class probability | 0.999900 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419233 120 + 20766785 GGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGCGCCAGUUAUCAAACGUAAGUGAAUU ..((((....))))((((((..(((((((.(((((((((..((((((..........))))))(((((...(((....))))))))..)))).))))).)))))))...))))))..... ( -44.90) >DroSec_CAF1 16790 120 + 1 GGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGAACCAGUUAUCAAGCGUAAGUGAAUU ........(.((((((((........(((((.((((....)))).))))).(((((((((..(.((......)).)..)).((((.....))))..)))))))..)))))))).)..... ( -41.10) >DroSim_CAF1 17477 120 + 1 GGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGCGCCAGUUAUCAAGCGUAAGUGAAUU ........(.(((((((.....(((((((.(((((((((..((((((..........))))))(((((...(((....))))))))..)))).))))).)))))))))))))).)..... ( -45.00) >DroEre_CAF1 13964 120 + 1 GGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUAGCAAUGCUAAAUGCAACGCCAGUUAUCAAGCGUAAGUGAAUU ........(.(((((((.....(((((((.((((.((((.(((((((..........)))))))..((.(((((....))))).))..))))..)))).)))))))))))))).)..... ( -39.90) >DroYak_CAF1 14434 120 + 1 GGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCAGUGCUAAAUGCAACGCCAGUUAUCAAACGUAAGUGAAUU ..((((....))))((((((..(((((((.((((.((((..(((((((((.(((((..((....))..))))).))))))..)))...))))..)))).)))))))...))))))..... ( -40.40) >consensus GGGCAAUAAGUUGCGCUUAUUAUGAUAAUGGGUGCGCAUGUGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGCGCCAGUUAUCAAGCGUAAGUGAAUU ..((((....))))((((((..(((((((.(((((((((..((((((..........))))))(((((...(((....))))))))..)))).))))).)))))))...))))))..... (-39.66 = -40.50 + 0.84)



| Location | 6,419,233 – 6,419,353 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 97.42 |
| Mean single sequence MFE | -31.68 |
| Consensus MFE | -31.99 |
| Energy contribution | -32.03 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -3.51 |
| Structure conservation index | 1.01 |
| SVM decision value | 5.23 |
| SVM RNA-class probability | 0.999980 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419233 120 - 20766785 AAUUCACUUACGUUUGAUAACUGGCGCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCC ...........((..(((...(((((((((.....))))))))).....)))..)).((((((..........))))))...(((((.................)))))........... ( -34.33) >DroSec_CAF1 16790 120 - 1 AAUUCACUUACGCUUGAUAACUGGUUCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCC ...........((..(((...((((.((((.....)))).)))).....)))..)).((((((..........))))))...(((((.................)))))........... ( -30.03) >DroSim_CAF1 17477 120 - 1 AAUUCACUUACGCUUGAUAACUGGCGCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCC ...........((..(((...(((((((((.....))))))))).....)))..)).((((((..........))))))...(((((.................)))))........... ( -35.93) >DroEre_CAF1 13964 120 - 1 AAUUCACUUACGCUUGAUAACUGGCGUUGCAUUUAGCAUUGCUACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCC ...........(((((((((.(((.(((((((.(((.((((......)))).)))..((((((..........))))))..))))).))))))))))).....)))((((....)))).. ( -30.10) >DroYak_CAF1 14434 120 - 1 AAUUCACUUACGUUUGAUAACUGGCGUUGCAUUUAGCACUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCC ...........((..(((...(((((.(((.....))).))))).....)))..)).((((((..........))))))...(((((.................)))))........... ( -28.03) >consensus AAUUCACUUACGCUUGAUAACUGGCGCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCACAUGCGCACCCAUUAUCAUAAUAAGCGCAACUUAUUGCCC ...........((..(((...(((((((((.....))))))))).....)))..)).((((((..........))))))...(((((.................)))))........... (-31.99 = -32.03 + 0.04)

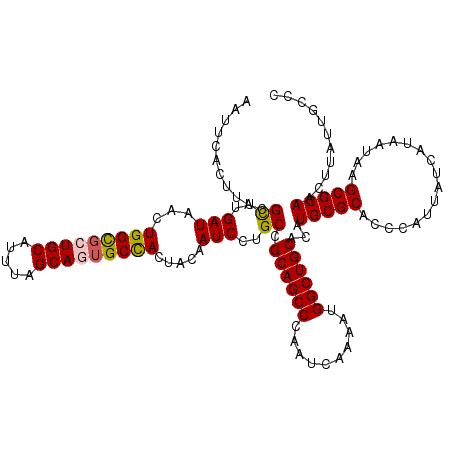
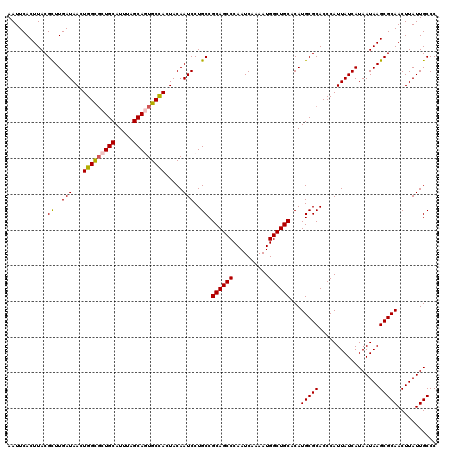
| Location | 6,419,273 – 6,419,393 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -43.84 |
| Consensus MFE | -40.56 |
| Energy contribution | -41.36 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.06 |
| Mean z-score | -3.23 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.85 |
| SVM RNA-class probability | 0.999956 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419273 120 + 20766785 UGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGCGCCAGUUAUCAAACGUAAGUGAAUUGAAUGUGCGCAGCUGCAUUGUCGGAAACCGGAAACGGAAA .((((((..........))))))(((((...(((((((((.((((.....)))).))).((..((((((....))...))))....))...))))))))))).....(((....)))... ( -44.10) >DroSec_CAF1 16830 120 + 1 UGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGAACCAGUUAUCAAGCGUAAGUGAAUUGAAUGUGCGCAGCUGCAUUGUCGGAAACCGGAAACGGAAA ((((((.....(((((((((..(.((......)).)..)).((((.....))))..)))))))....(((((.((.......)).))))).))))))..........(((....)))... ( -43.20) >DroSim_CAF1 17517 120 + 1 UGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGCGCCAGUUAUCAAGCGUAAGUGAAUUGAAUGUGCGCAGCUGCAUUGUCGGAAACCGGAAACGGAAA .((((((..........))))))(((((...(((((((((.((((.....)))).))).........(((((.((.......)).))))).))))))))))).....(((....)))... ( -47.80) >DroEre_CAF1 14004 120 + 1 UGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUAGCAAUGCUAAAUGCAACGCCAGUUAUCAAGCGUAAGUGAAUUGAAUGUGCGCAGCUGCACUGUUGGCCACCGGAAACGGAAA .((((((..........))))))(((..(((.((.(((((..(((.....))).(((((.(((....((....))....))).)).)))..))))))).))).))).(((....)))... ( -44.10) >DroYak_CAF1 14474 120 + 1 UGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCAGUGCUAAAUGCAACGCCAGUUAUCAAACGUAAGUGAAUUGAAUGUGCGCAGCUGCAUUGUUGAAAACCGGAAACGGAAA .((((((..........))))))..(((.(.((((((.((..(((.....)))..((((.(((....((....))....))).)).)))).)))))).).)))....(((....)))... ( -40.00) >consensus UGCAGCCAUUUUGAUUGGGCUGCGGCAGGAUUGUAGUGGCACUGCUAAAUGCAGCGCCAGUUAUCAAGCGUAAGUGAAUUGAAUGUGCGCAGCUGCAUUGUCGGAAACCGGAAACGGAAA .((((((..........))))))(((((...(((((((((.((((.....)))).))).........(((((.((.......)).))))).))))))))))).....(((....)))... (-40.56 = -41.36 + 0.80)

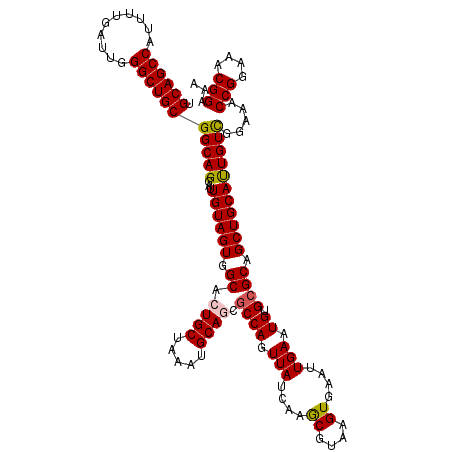
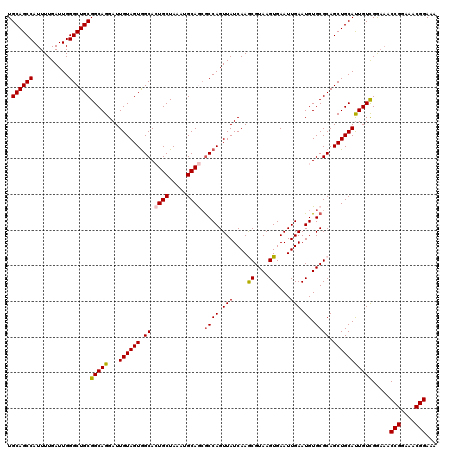
| Location | 6,419,273 – 6,419,393 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.58 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -30.01 |
| Energy contribution | -30.05 |
| Covariance contribution | 0.04 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.84 |
| SVM RNA-class probability | 0.997317 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6419273 120 - 20766785 UUUCCGUUUCCGGUUUCCGACAAUGCAGCUGCGCACAUUCAAUUCACUUACGUUUGAUAACUGGCGCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCA ...(((....)))..........((((((((((((...((((...........))))....(((((((((.....))))))))).........)).))))))(((.......))).)))) ( -33.60) >DroSec_CAF1 16830 120 - 1 UUUCCGUUUCCGGUUUCCGACAAUGCAGCUGCGCACAUUCAAUUCACUUACGCUUGAUAACUGGUUCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCA ...(((....)))...........((((..(((.................)))........((((.((((.....)))).))))........)))).((((((..........)))))). ( -28.33) >DroSim_CAF1 17517 120 - 1 UUUCCGUUUCCGGUUUCCGACAAUGCAGCUGCGCACAUUCAAUUCACUUACGCUUGAUAACUGGCGCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCA ...(((....)))...........((((..(((.................)))........(((((((((.....)))))))))........)))).((((((..........)))))). ( -34.23) >DroEre_CAF1 14004 120 - 1 UUUCCGUUUCCGGUGGCCAACAGUGCAGCUGCGCACAUUCAAUUCACUUACGCUUGAUAACUGGCGUUGCAUUUAGCAUUGCUACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCA ............(..((((...((((......)))).................(((((...(((.(((((...(((.((((......)))).)))..)))))))))))))..))))..). ( -31.60) >DroYak_CAF1 14474 120 - 1 UUUCCGUUUCCGGUUUUCAACAAUGCAGCUGCGCACAUUCAAUUCACUUACGUUUGAUAACUGGCGUUGCAUUUAGCACUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCA ...........(((.......((((((((.((.((...((((...........))))....)))))))))))).......)))..............((((((..........)))))). ( -27.64) >consensus UUUCCGUUUCCGGUUUCCGACAAUGCAGCUGCGCACAUUCAAUUCACUUACGCUUGAUAACUGGCGCUGCAUUUAGCAGUGCCACUACAAUCCUGCCGCAGCCCAAUCAAAAUGGCUGCA ...(((....)))...........((((..(((.................)))........(((((((((.....)))))))))........)))).((((((..........)))))). (-30.01 = -30.05 + 0.04)

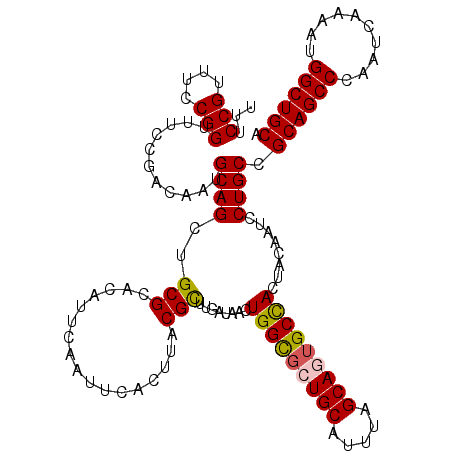
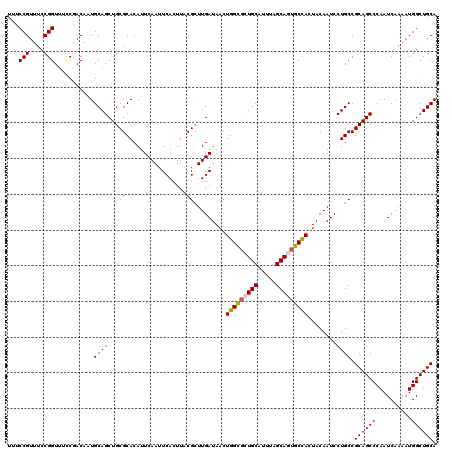
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:59 2006