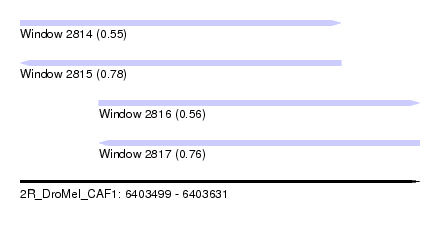
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,403,499 – 6,403,631 |
| Length | 132 |
| Max. P | 0.784219 |

| Location | 6,403,499 – 6,403,605 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -30.59 |
| Consensus MFE | -18.92 |
| Energy contribution | -20.35 |
| Covariance contribution | 1.42 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.549078 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
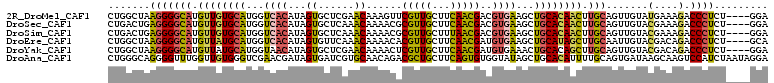
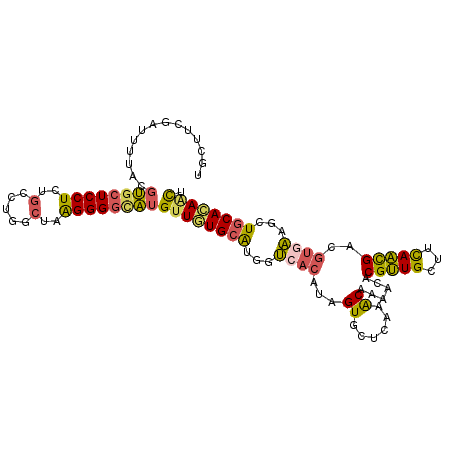
>2R_DroMel_CAF1 6403499 106 + 20766785 UCC----AGAGGGUCUUUCAUACAACUGCAAGUUGUGCAGCUUCACGUCGUUGAAGCAACGAACUUUGUUCGAGCACUAUGUGACCAUGCACAACAUGCCCCUUAGCCAG ...----.(((((..............(((.((((((((((((((......))))))..(((((...)))))...............)))))))).))))))))...... ( -32.03) >DroSec_CAF1 756 106 + 1 UCC----AGAGGGUCUUUCGUACAACUGCAAGUUGUGCAGCUUCACGUCGUUGAAGCAACGCGUUUUGUUUGAGCACUAUGUGACCAUGCACAACAUGCCCCUCAGUCAG ...----.(((((......(.....).(((.((((((((((((((......))))))..(((((..(((....)))..)))))....)))))))).))))))))...... ( -37.20) >DroSim_CAF1 670 106 + 1 UCC----AGAGGGUCUUUCGUACAACUGCAAGUUGUGCAGCUUCACGUCGUUAAAGCAACGCGUUUUGUUUGAGCACUAUGUGACCAUGCACAACAUGCCCCUCAGUCAG ...----.(((((......(.....).(((.((((((((...((((((.(((.((((((......)))))).)))...))))))...)))))))).))))))))...... ( -34.30) >DroEre_CAF1 740 106 + 1 UGC----AGAGGGUCUGUCGUACAAUUGCAAGCUAUGCAGCUUCACAUCGUUGAAGCAACGUGUUUUGUUUGAACACUAUGUGACCAUGCAUAACAUGCCCCUUAGCCAG .((----.(((((..((.....))...(((.(.((((((...((((((.((((...))))((((((.....)))))).))))))...)))))).).)))))))).))... ( -32.00) >DroYak_CAF1 671 106 + 1 UCC----AGAGGGUCUGUCGUACAACUGCAAGCUGUGCAGUUUCACAUCGUUGAAGCAACGAGUUUUGUUCGAGCACUAUGUUACCAUGCAUAACAUGCCCCUUAGCCAG ...----.(((((....(((.(((((((((.....))))))......((((((...))))))....))).)))(((.(((((......)))))...))))))))...... ( -27.40) >DroAna_CAF1 620 110 + 1 UCCUAUUAGAUGGACUUGCUUAUCACUGCAAAAUGUGCAGCUAUACCACACUGAAGCAGCGUCUGUUGCACGAUCACUAUCGUUCGACCCACAACCAAACCCCUGCCCAG ......(((..((((((((((....(((((.....)))))......(.....))))))).))))((((.(((((....))))).))))..............)))..... ( -20.60) >consensus UCC____AGAGGGUCUUUCGUACAACUGCAAGUUGUGCAGCUUCACGUCGUUGAAGCAACGAGUUUUGUUCGAGCACUAUGUGACCAUGCACAACAUGCCCCUUAGCCAG ........(((((..............(((.(.((((((...((((((.(((..(((((......)))))..)))...))))))...)))))).).))))))))...... (-18.92 = -20.35 + 1.42)


| Location | 6,403,499 – 6,403,605 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 81.37 |
| Mean single sequence MFE | -35.20 |
| Consensus MFE | -23.05 |
| Energy contribution | -22.87 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.44 |
| Mean z-score | -2.07 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.784219 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6403499 106 - 20766785 CUGGCUAAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCGAACAAAGUUCGUUGCUUCAACGACGUGAAGCUGCACAACUUGCAGUUGUAUGAAAGACCCUCU----GGA (..(...(((((((.((((((((...((((...((......))...((.(((((...)))))))))))...)))))))).))).......(....).))))).----.). ( -32.90) >DroSec_CAF1 756 106 - 1 CUGACUGAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCAAACAAAACGCGUUGCUUCAACGACGUGAAGCUGCACAACUUGCAGUUGUACGAAAGACCCUCU----GGA ....(.((((((((.((((((((((..(((...)))..)).......(((((((......)))))))....)))))))).))).......(....).))))).----).. ( -37.90) >DroSim_CAF1 670 106 - 1 CUGACUGAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCAAACAAAACGCGUUGCUUUAACGACGUGAAGCUGCACAACUUGCAGUUGUACGAAAGACCCUCU----GGA ....(.((((((((.((((((((((..(((...)))..)).......(((((((......)))))))....)))))))).))).......(....).))))).----).. ( -37.90) >DroEre_CAF1 740 106 - 1 CUGGCUAAGGGGCAUGUUAUGCAUGGUCACAUAGUGUUCAAACAAAACACGUUGCUUCAACGAUGUGAAGCUGCAUAGCUUGCAAUUGUACGACAGACCCUCU----GCA ...((..(((((((.((((((((...((((((.(((((.......)))))((((...)))).))))))...)))))))).)))..(((.....))).))))..----)). ( -37.80) >DroYak_CAF1 671 106 - 1 CUGGCUAAGGGGCAUGUUAUGCAUGGUAACAUAGUGCUCGAACAAAACUCGUUGCUUCAACGAUGUGAAACUGCACAGCUUGCAGUUGUACGACAGACCCUCU----GGA ....(((..(((..(((..(((.......((((((......)).....((((((...)))))))))).(((((((.....))))))))))..)))..)))..)----)). ( -31.00) >DroAna_CAF1 620 110 - 1 CUGGGCAGGGGUUUGGUUGUGGGUCGAACGAUAGUGAUCGUGCAACAGACGCUGCUUCAGUGUGGUAUAGCUGCACAUUUUGCAGUGAUAAGCAAGUCCAUCUAAUAGGA .(((((....((((((((((((((((........))))).))))))...((((((....(((..(.....)..))).....)))))).)))))..))))).......... ( -33.70) >consensus CUGGCUAAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCAAACAAAACACGUUGCUUCAACGACGUGAAGCUGCACAACUUGCAGUUGUACGAAAGACCCUCU____GGA .......(((((((.((((((((...((((...((......))......(((((...)))))..))))...)))))))).))).......(....).))))......... (-23.05 = -22.87 + -0.19)


| Location | 6,403,525 – 6,403,631 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -19.62 |
| Energy contribution | -20.43 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.562129 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6403525 106 + 20766785 AGUUGUGCAGCUUCACGUCGUUGAAGCAACGAACUUUGUUCGAGCACUAUGUGACCAUGCACAACAUGCCCCUUAGCCAGGCAGAGGAGUACGUAAAAUCAAAGCA .((((((((((((((......))))))..(((((...)))))...............)))))))).(((.((((.((...)).)))).)))............... ( -31.10) >DroSec_CAF1 782 106 + 1 AGUUGUGCAGCUUCACGUCGUUGAAGCAACGCGUUUUGUUUGAGCACUAUGUGACCAUGCACAACAUGCCCCUCAGUCAGGCAGAGGAGCACGUAAAAUCGAAGCA .((((((((((((((......))))))..(((((..(((....)))..)))))....)))))))).(((.((((.(.....).)))).)))((......))..... ( -35.70) >DroSim_CAF1 696 106 + 1 AGUUGUGCAGCUUCACGUCGUUAAAGCAACGCGUUUUGUUUGAGCACUAUGUGACCAUGCACAACAUGCCCCUCAGUCAGGCAGAGGAGCACGUAAAAUCGAAGCA .((((((((...((((((.(((.((((((......)))))).)))...))))))...)))))))).(((.((((.(.....).)))).)))((......))..... ( -32.80) >DroEre_CAF1 766 106 + 1 AGCUAUGCAGCUUCACAUCGUUGAAGCAACGUGUUUUGUUUGAACACUAUGUGACCAUGCAUAACAUGCCCCUUAGCCAGGCAGAGGAGCACGUAAAAUCAAAGCA .((((((((...((((((.((((...))))((((((.....)))))).))))))...))))))((.(((.((((.((...)).)))).))).)).........)). ( -32.30) >DroYak_CAF1 697 106 + 1 AGCUGUGCAGUUUCACAUCGUUGAAGCAACGAGUUUUGUUCGAGCACUAUGUUACCAUGCAUAACAUGCCCCUUAGCCAGGCAGAGGAGCACGUUAAAUCAAAGCA .(((((((.((((((......))))))..((((.....)))).))))..............((((.(((.((((.((...)).)))).))).))))......))). ( -28.90) >DroAna_CAF1 650 106 + 1 AAAUGUGCAGCUAUACCACACUGAAGCAGCGUCUGUUGCACGAUCACUAUCGUUCGACCCACAACCAAACCCCUGCCCAGGCAGAGGAGCUCUCUAAGGUGAAGCU ........((((.((((.....(((((...((..((((.(((((....))))).))))..)).......((.((((....)))).)).))).))...)))).)))) ( -29.80) >consensus AGUUGUGCAGCUUCACGUCGUUGAAGCAACGAGUUUUGUUCGAGCACUAUGUGACCAUGCACAACAUGCCCCUUAGCCAGGCAGAGGAGCACGUAAAAUCAAAGCA ...((((((...((((((.(((..(((((......)))))..)))...))))))...))))))((.(((.((((.((...)).)))).))).))............ (-19.62 = -20.43 + 0.81)



| Location | 6,403,525 – 6,403,631 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 82.08 |
| Mean single sequence MFE | -34.77 |
| Consensus MFE | -24.61 |
| Energy contribution | -24.28 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.43 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.71 |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.756274 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6403525 106 - 20766785 UGCUUUGAUUUUACGUACUCCUCUGCCUGGCUAAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCGAACAAAGUUCGUUGCUUCAACGACGUGAAGCUGCACAACU .................(((((..((...))..)))))...((((((((...((((...((......))...((.(((((...)))))))))))...)))))))). ( -29.60) >DroSec_CAF1 782 106 - 1 UGCUUCGAUUUUACGUGCUCCUCUGCCUGACUGAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCAAACAAAACGCGUUGCUUCAACGACGUGAAGCUGCACAACU ..............(((((((((.........)))))))))((((((((((..(((...)))..)).......(((((((......)))))))....)))))))). ( -36.50) >DroSim_CAF1 696 106 - 1 UGCUUCGAUUUUACGUGCUCCUCUGCCUGACUGAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCAAACAAAACGCGUUGCUUUAACGACGUGAAGCUGCACAACU ..............(((((((((.........)))))))))((((((((((..(((...)))..)).......(((((((......)))))))....)))))))). ( -36.50) >DroEre_CAF1 766 106 - 1 UGCUUUGAUUUUACGUGCUCCUCUGCCUGGCUAAGGGGCAUGUUAUGCAUGGUCACAUAGUGUUCAAACAAAACACGUUGCUUCAACGAUGUGAAGCUGCAUAGCU .((.........((((((((((..((...))..))))))))))((((((...((((((.(((((.......)))))((((...)))).))))))...)))))))). ( -36.90) >DroYak_CAF1 697 106 - 1 UGCUUUGAUUUAACGUGCUCCUCUGCCUGGCUAAGGGGCAUGUUAUGCAUGGUAACAUAGUGCUCGAACAAAACUCGUUGCUUCAACGAUGUGAAACUGCACAGCU ((((.((...((((((((((((..((...))..))))))))))))..)).)))).....((((....(((....((((((...)))))))))......)))).... ( -35.00) >DroAna_CAF1 650 106 - 1 AGCUUCACCUUAGAGAGCUCCUCUGCCUGGGCAGGGGUUUGGUUGUGGGUCGAACGAUAGUGAUCGUGCAACAGACGCUGCUUCAGUGUGGUAUAGCUGCACAUUU ((((..(((..........(((((((....)))))))....(((((((((((........))))).))))))..((((((...)))))))))..))))........ ( -34.10) >consensus UGCUUCGAUUUUACGUGCUCCUCUGCCUGGCUAAGGGGCAUGUUGUGCAUGGUCACAUAGUGCUCAAACAAAACACGUUGCUUCAACGACGUGAAGCUGCACAACU ..............((((((((..(.....)..))))))))((((((((...((((...((......))......(((((...)))))..))))...)))))))). (-24.61 = -24.28 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:43 2006