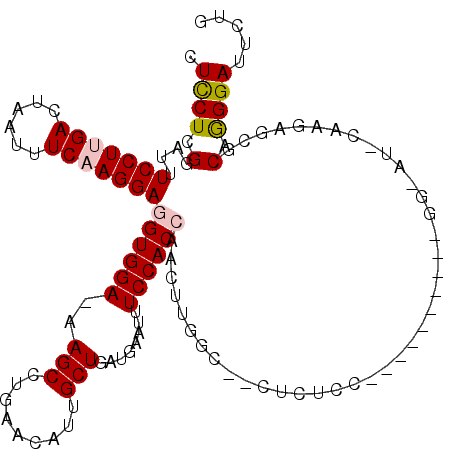
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,377,944 – 6,378,062 |
| Length | 118 |
| Max. P | 0.783484 |

| Location | 6,377,944 – 6,378,062 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 77.90 |
| Mean single sequence MFE | -32.14 |
| Consensus MFE | -17.55 |
| Energy contribution | -17.71 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.55 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518803 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6377944 118 + 20766785 CAGAAUCCCUGCGCUCUUGGAUACCAAGUGCUUUUGGAGAGCAGCCAAGUUGGUGGAAAUUCAUCAGCAAUGUUCAGGCUU-UCCACCUCCUUGAAAUUAGUCAAGGAAAUCCGAGAAG (((.....)))...((((((((............((((((((.((...((((((((....))))))))...))....))))-))))..(((((((......))))))).)))))))).. ( -38.70) >DroSec_CAF1 18679 101 + 1 CAGAAUCCCUGCGCUCUU-----------------GGAGAGUGGCCAAGUUGGUGGAAAUUCAUCAGCACUGCUCAGGCUU-UCCACCUCCUUGAAAUUAGUCAAGGAAAUCCGAGGAG .......(((.((....(-----------------((((((((((...((((((((....))))))))...)))...))))-))))..(((((((......)))))))....))))).. ( -32.60) >DroSim_CAF1 19951 101 + 1 CAGAAUCCCUGCGCUCUU-----------------GGAGAGUGGCCAAGUUGGUGGAAAUUCAUAAGCACUGUUCAGGCUU-UCCACCUCCUUGAAAUUAGUCAAGGAAAUCCGAGGAG (((.....)))...((((-----------------(((.............(((((((.......(((.((....))))))-))))))(((((((......)))))))..))))))).. ( -29.91) >DroEre_CAF1 19849 97 + 1 CAGAAUCCUUGUACUCUGGGAUCCC-U--------GGAGA------------GUGGAAAUUU-UCAGCAAUGUUCAGGCUUUUCCACCUCCUUGAAAUUAGUCGAGGAAAUCCAAGGAG .....((((((..(((..((...))-.--------.))).------------(((((((...-..(((.........)))))))))).(((((((......)))))))....)))))). ( -31.70) >DroYak_CAF1 19896 105 + 1 CAGAAUCCCUGUACUCUUGCAUCCC-UGUGCUCGUGGAGA------------GUGGAAAUUU-UCAGCAAUGUUCAGGCUUUUCCACCUCCUUGAAAUUAGUCCAGGAAAUCCGAGGAG .......((((.(((...((((...-.))))(((.((((.------------(((((((...-..(((.........)))))))))))))).)))....))).))))...((....)). ( -27.80) >consensus CAGAAUCCCUGCGCUCUUG_AU_CC__________GGAGAG__GCCAAGUUGGUGGAAAUUCAUCAGCAAUGUUCAGGCUU_UCCACCUCCUUGAAAUUAGUCAAGGAAAUCCGAGGAG (((.....)))........................(((.............((((((........(((...)))........))))))(((((((......)))))))..)))...... (-17.55 = -17.71 + 0.16)

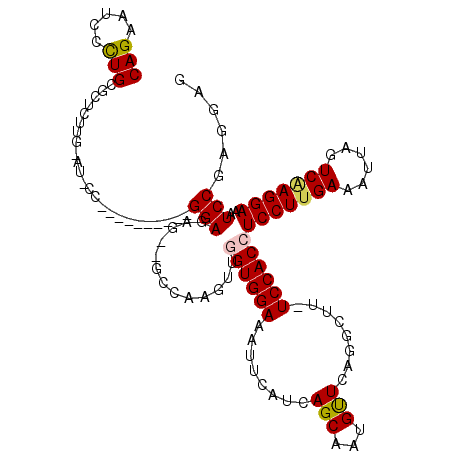
| Location | 6,377,944 – 6,378,062 |
|---|---|
| Length | 118 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 77.90 |
| Mean single sequence MFE | -32.27 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.86 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783484 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6377944 118 - 20766785 CUUCUCGGAUUUCCUUGACUAAUUUCAAGGAGGUGGA-AAGCCUGAACAUUGCUGAUGAAUUUCCACCAACUUGGCUGCUCUCCAAAAGCACUUGGUAUCCAAGAGCGCAGGGAUUCUG .....((((..((((((........((((..((((((-((..(..............)..))))))))..))))((((((.......))).(((((...)))))))).)))))).)))) ( -35.94) >DroSec_CAF1 18679 101 - 1 CUCCUCGGAUUUCCUUGACUAAUUUCAAGGAGGUGGA-AAGCCUGAGCAGUGCUGAUGAAUUUCCACCAACUUGGCCACUCUCC-----------------AAGAGCGCAGGGAUUCUG .((((((....(((((((......)))))))((((((-((..((.(((...))).).)..))))))))..(((((.......))-----------------)))..)).))))...... ( -32.80) >DroSim_CAF1 19951 101 - 1 CUCCUCGGAUUUCCUUGACUAAUUUCAAGGAGGUGGA-AAGCCUGAACAGUGCUUAUGAAUUUCCACCAACUUGGCCACUCUCC-----------------AAGAGCGCAGGGAUUCUG .((((((....(((((((......)))))))((((((-((((((....)).))).......)))))))..(((((.......))-----------------)))..)).))))...... ( -31.11) >DroEre_CAF1 19849 97 - 1 CUCCUUGGAUUUCCUCGACUAAUUUCAAGGAGGUGGAAAAGCCUGAACAUUGCUGA-AAAUUUCCAC------------UCUCC--------A-GGGAUCCCAGAGUACAAGGAUUCUG .((((((((..((((.((......)).))))(((((((((((.........)))..-...)))))))------------).)))--------)-))))...((((((......)))))) ( -29.20) >DroYak_CAF1 19896 105 - 1 CUCCUCGGAUUUCCUGGACUAAUUUCAAGGAGGUGGAAAAGCCUGAACAUUGCUGA-AAAUUUCCAC------------UCUCCACGAGCACA-GGGAUGCAAGAGUACAGGGAUUCUG .....((((.((((((.(((........((((((((((((((.........)))..-...)))))))------------.))))....(((..-....)))...))).)))))).)))) ( -32.30) >consensus CUCCUCGGAUUUCCUUGACUAAUUUCAAGGAGGUGGA_AAGCCUGAACAUUGCUGAUGAAUUUCCACCAACUUGGC__CUCUCC__________GG_AU_CAAGAGCGCAGGGAUUCUG .((((.(....(((((((......)))))))((((((..(((.........)))........))))))........................................).))))..... (-17.38 = -17.86 + 0.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:34 2006