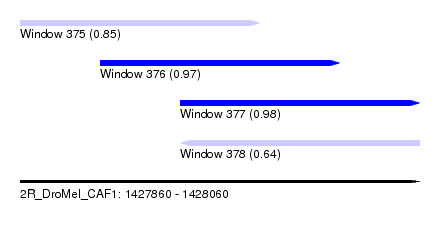
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,427,860 – 1,428,060 |
| Length | 200 |
| Max. P | 0.978774 |

| Location | 1,427,860 – 1,427,980 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 98.33 |
| Mean single sequence MFE | -39.48 |
| Consensus MFE | -35.88 |
| Energy contribution | -36.68 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850877 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1427860 120 + 20766785 CAAUGGCAUUACUUCACCCACUCGCCUCAUGUGGCUGCUCGUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUG ...............((((....(((......)))((((.((....)).))))........(((((......)))))))))..(((((((((((((..........))))))).)))))) ( -40.50) >DroSec_CAF1 46314 120 + 1 CAAUGGCAUUACUUCACCCACUCGCCUCAUGUGGCUGCUCGUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUG ...............((((....(((......)))((((.((....)).))))........(((((......)))))))))..(((((((((((((..........))))))).)))))) ( -40.50) >DroSim_CAF1 51364 120 + 1 CAAUGGCAUUACUUCACCCACUCGCCUCAUGUGGCUGCUCGUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUG ...............((((....(((......)))((((.((....)).))))........(((((......)))))))))..(((((((((((((..........))))))).)))))) ( -40.50) >DroEre_CAF1 44446 120 + 1 CAAUGGCAUUACUUCACCCACUCGCCUCAUGUGGCUGCUCGUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCAGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUG ...((((((..............(((......))).((..((.(((((....))))).))..))((....)))))))).....(((((((((((((..........))))))).)))))) ( -38.10) >DroYak_CAF1 48966 120 + 1 CAAUGGCAUUACUUCACCCACUCGACUCAUGUGGCUGCUCGUUAUUAUUAGCAUAAUUACAGGCGCAUUAACGUGCCGGGUUUCAGGGGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUA ....(((.........(((.((.(((((.(((((.((((.((....)).))))...)))))(((((......))))))))))..)))))(((((((..........)))))))..))).. ( -37.80) >consensus CAAUGGCAUUACUUCACCCACUCGCCUCAUGUGGCUGCUCGUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUG ...............((((....(((......)))((((.((....)).))))........(((((......)))))))))..(((((((((((((..........))))))).)))))) (-35.88 = -36.68 + 0.80)

| Location | 1,427,900 – 1,428,020 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 96.50 |
| Mean single sequence MFE | -40.20 |
| Consensus MFE | -35.46 |
| Energy contribution | -35.70 |
| Covariance contribution | 0.24 |
| Combinations/Pair | 1.03 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.69 |
| SVM RNA-class probability | 0.972185 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1427900 120 + 20766785 GUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGACAGGGCCUAAAAGCGAGACGCAUCCACAAUGCA ..........((((.......(((((......)))))((((.((((((((((((((..........))))))).)))))))..................(((...)))))))...)))). ( -39.80) >DroSec_CAF1 46354 120 + 1 GUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGGGACGCAUCCACAAUGCA ..........((((.......(((((......)))))((((.((((((((((((((..........))))))).)))))))........(.((((......))).).)))))...)))). ( -41.30) >DroSim_CAF1 51404 120 + 1 GUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGGGACGCAUCCACAAUGCA ..........((((.......(((((......)))))((((.((((((((((((((..........))))))).)))))))........(.((((......))).).)))))...)))). ( -41.30) >DroEre_CAF1 44486 120 + 1 GUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCAGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGAGACGCAUCCACAAUGCA ..........((((.......(((((......)))))(((((((((((((((((((..........))))))).))))))).........)))))....(((...))).......)))). ( -38.70) >DroYak_CAF1 49006 120 + 1 GUUAUUAUUAGCAUAAUUACAGGCGCAUUAACGUGCCGGGUUUCAGGGGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUAAUUAUGAUAGGGCCUUAAAACGAGGCGCAUCUACAAUGCA .......(((.(((((((...(((((......)))))((((.((...(((..(((.....)))....)))..)).))))))))))).))).(((((.....)))))((((.....)))). ( -39.90) >consensus GUUAUUAUUAGCAUAAUUACAGGCGCAUUAGCGUGCCGGGUUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGAGACGCAUCCACAAUGCA ((((....)))).........(((((......)))))(((((((((((((((((((..........))))))).))))))).........)))))...........((((.....)))). (-35.46 = -35.70 + 0.24)

| Location | 1,427,940 – 1,428,060 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -43.82 |
| Consensus MFE | -36.86 |
| Energy contribution | -37.14 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.84 |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.978774 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1427940 120 + 20766785 UUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGACAGGGCCUAAAAGCGAGACGCAUCCACAAUGCACCUCCUGCAGUGCCGCUUAAGCGCUUAAAGUUGCGCAGUU ..((((((((((((((..........))))))).)))))))....(((..(((((...((.(((..((((.....))))..)))))..)).)))......((((........)))).))) ( -42.90) >DroSec_CAF1 46394 120 + 1 UUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGGGACGCAUCCACAAUGCACCUCCUGCAGUGCCGCUUAAGCGCUUAAAGUUGCGCAGUU ..((((((((((((((..........))))))).))))))).........(((((....((((((.((((.....))))...)))))))).)))......((((........)))).... ( -45.10) >DroSim_CAF1 51444 120 + 1 UUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGGGACGCAUCCACAAUGCACCUCCUGCAGUGCCGCUUAAGCGCUUAAAGUUGCGCAGUU ..((((((((((((((..........))))))).))))))).........(((((....((((((.((((.....))))...)))))))).)))......((((........)))).... ( -45.10) >DroEre_CAF1 44526 120 + 1 UUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGAGACGCAUCCACAAUGCACCUCCUGCAGCGCCGCUGGAGCGUUUAAGGUUUCGCAGUU ..((((((((((((((..........))))))).)))))))..................(((((((((((.....))))((((((.((......)).)))).)).....))))))).... ( -44.30) >DroYak_CAF1 49046 120 + 1 UUUCAGGGGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUAAUUAUGAUAGGGCCUUAAAACGAGGCGCAUCUACAAUGCACCUCCUGCAGCGCUGCUAAAGCGUUUAAGGUUGCGCAGUU ....((.(((..(((.....))).(((((.((((..((((.......))))(((((.....)))))((((.....))))...)))))))))))).))...((((........)))).... ( -41.70) >consensus UUUCAGGUGCUUGGCAAUUUGCCUGCUGCCAGGACGCCUGAUUAUGAUAGGGCCUAAAAGCGAGACGCAUCCACAAUGCACCUCCUGCAGUGCCGCUUAAGCGCUUAAAGUUGCGCAGUU ..((((((((((((((..........))))))).))))))).........(((((...((.(((..((((.....))))..)))))..)).)))......((((........)))).... (-36.86 = -37.14 + 0.28)

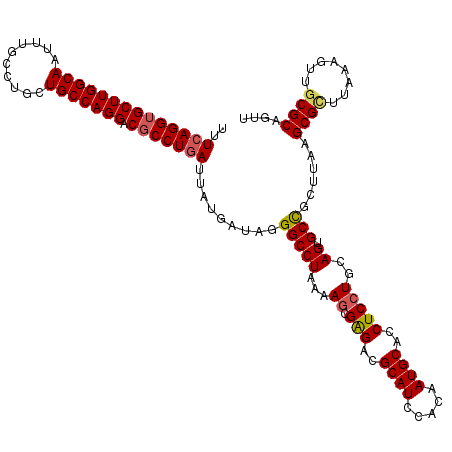
| Location | 1,427,940 – 1,428,060 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 94.08 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -33.34 |
| Energy contribution | -34.06 |
| Covariance contribution | 0.72 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643092 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1427940 120 - 20766785 AACUGCGCAACUUUAAGCGCUUAAGCGGCACUGCAGGAGGUGCAUUGUGGAUGCGUCUCGCUUUUAGGCCCUGUCAUAAUCAGGCGUCCUGGCAGCAGGCAAAUUGCCAAGCACCUGAAA ..((((((........))(((.....)))...)))).((((((.(((..(((..((((.(((.((((((((((.......)))).).))))).)))))))..)))..))))))))).... ( -42.40) >DroSec_CAF1 46394 120 - 1 AACUGCGCAACUUUAAGCGCUUAAGCGGCACUGCAGGAGGUGCAUUGUGGAUGCGUCCCGCUUUUAGGCCCUAUCAUAAUCAGGCGUCCUGGCAGCAGGCAAAUUGCCAAGCACCUGAAA ....((((........))))...((.(((.(((.(((.((((((((...)))))).))..))).)))))))).......(((((.((..((((((........)))))).)).))))).. ( -38.40) >DroSim_CAF1 51444 120 - 1 AACUGCGCAACUUUAAGCGCUUAAGCGGCACUGCAGGAGGUGCAUUGUGGAUGCGUCCCGCUUUUAGGCCCUAUCAUAAUCAGGCGUCCUGGCAGCAGGCAAAUUGCCAAGCACCUGAAA ....((((........))))...((.(((.(((.(((.((((((((...)))))).))..))).)))))))).......(((((.((..((((((........)))))).)).))))).. ( -38.40) >DroEre_CAF1 44526 120 - 1 AACUGCGAAACCUUAAACGCUCCAGCGGCGCUGCAGGAGGUGCAUUGUGGAUGCGUCUCGCUUUUAGGCCCUAUCAUAAUCAGGCGUCCUGGCAGCAGGCAAAUUGCCAAGCACCUGAAA ...(((............(((((((.((((((....(((..(((((...)))))..)))(((....))).............)))))))))).))).(((.....)))..)))....... ( -40.50) >DroYak_CAF1 49046 120 - 1 AACUGCGCAACCUUAAACGCUUUAGCAGCGCUGCAGGAGGUGCAUUGUAGAUGCGCCUCGUUUUAAGGCCCUAUCAUAAUUAGGCGUCCUGGCAGCAGGCAAAUUGCCAAGCCCCUGAAA ....(((..........)))(((((..(((((((..((((((((((...))))))))))......((((((((.......)))).).))).))))).(((.....)))..))..))))). ( -43.90) >consensus AACUGCGCAACUUUAAGCGCUUAAGCGGCACUGCAGGAGGUGCAUUGUGGAUGCGUCUCGCUUUUAGGCCCUAUCAUAAUCAGGCGUCCUGGCAGCAGGCAAAUUGCCAAGCACCUGAAA ..((((((........))(((.....)))...)))).((((((.(((..(((..((((.(((.(((((((((.........))).).))))).)))))))..)))..))))))))).... (-33.34 = -34.06 + 0.72)

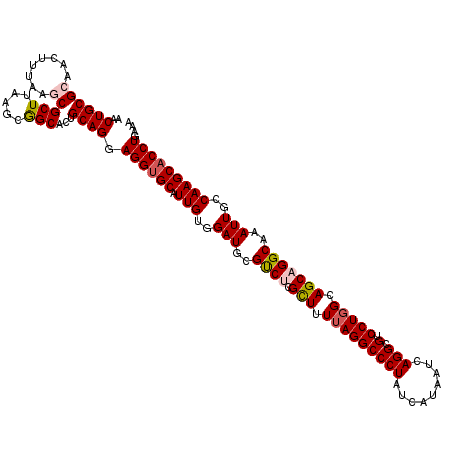
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:55 2006