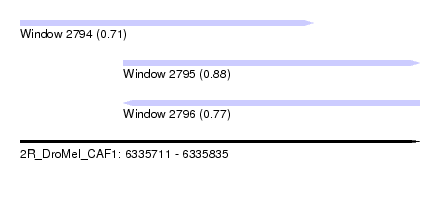
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,335,711 – 6,335,835 |
| Length | 124 |
| Max. P | 0.875957 |

| Location | 6,335,711 – 6,335,802 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 88.67 |
| Mean single sequence MFE | -20.23 |
| Consensus MFE | -18.87 |
| Energy contribution | -19.03 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.25 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.36 |
| SVM RNA-class probability | 0.705213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6335711 91 + 20766785 UUUGGCCAUUGAUGGAUUGAUGGAACGCCCAG--AGUUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGAAAGCCAAUCACAGGCA ...((((.......((((..(((.....))).--))))....((((.....))))))))..............---......(((.......))). ( -21.00) >DroPse_CAF1 1015 87 + 1 UUUUGCCUACG----UUAUAUGGAACGCCCAGGGAGAUAAUUUGGCU--AAGCCAGGCCUAAA-AGAAACUACAAGAAGAAAGCCAUUC--GGGCA ..........(----((......)))((((.(((......((((((.--..)))))))))...-...........(((........)))--)))). ( -15.60) >DroSec_CAF1 5788 87 + 1 UUUGGCCAACG----AUUGAUGGAACGCCCAG--AGUUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGAAAGCCAAUCACAGGCA ...((((...(----(((..(((.....))).--))))....((((.....))))))))..............---......(((.......))). ( -20.90) >DroSim_CAF1 5795 87 + 1 UUUGGCCAACG----AUUGAUGGAACGCCCAG--AGUUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGAAAGCCAAUCACAGGCA ...((((...(----(((..(((.....))).--))))....((((.....))))))))..............---......(((.......))). ( -20.90) >DroEre_CAF1 5770 87 + 1 UUUGGCCAACG----AUUAGUGGAACGCCCAG--AGCUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGAAAGCCAAUCACAGGCA ...((((...(----((((((((.....))..--.)))))))((((.....))))))))..............---......(((.......))). ( -22.70) >DroYak_CAF1 5738 87 + 1 UUUGGCCAACG----AUUAGUGGAACGCCCAG--AUUUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGAAAGCCAAUCACAGGCA ...((((...(----((((((((.....))).--..))))))((((.....))))))))..............---......(((.......))). ( -20.30) >consensus UUUGGCCAACG____AUUGAUGGAACGCCCAG__AGUUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA___AAGAAAGCCAAUCACAGGCA ...((((.............(((.....)))...........((((.....)))))))).......................(((.......))). (-18.87 = -19.03 + 0.17)

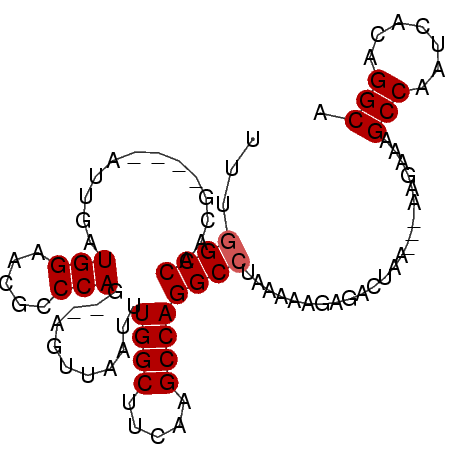
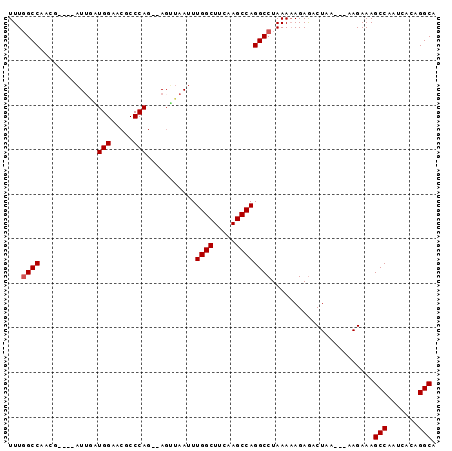
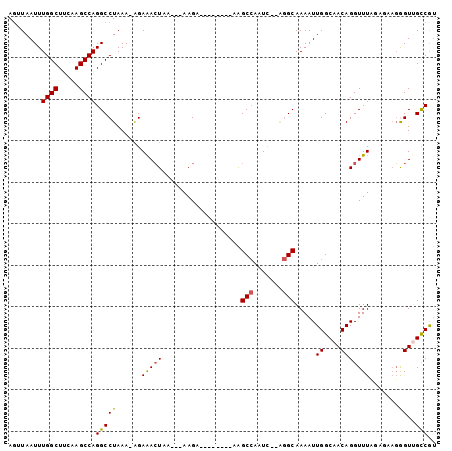
| Location | 6,335,743 – 6,335,835 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -16.48 |
| Energy contribution | -16.20 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.89 |
| SVM RNA-class probability | 0.875957 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6335743 92 + 20766785 AGUUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGA--------AAGCCAAUCACAGGCAAAAUUGGCAACAGGUUUCGAGCUGGGUUGCCGU ........((((.....))))((((((....((((((..---....--------..(((.......))).....((....))))))))....))))))..... ( -24.00) >DroPse_CAF1 1045 90 + 1 AGAUAAUUUGGCU--AAGCCAGGCCUAAA-AGAAACUACAAGAAGA--------AAGCCAUUC--GGGCAAAAUUGGCAACAGGUUUAUCCAAGAGUGGCCAU .........((((--......))))....-................--------..(((((((--((..(((.(((....))).)))..))..)))))))... ( -22.50) >DroGri_CAF1 5368 95 + 1 AGUUAAUUUGGCUUUAAGCCAGGCCUAAA-GGAAAAUAG---AAGAGAAAAUUGAGGCGAAUU----GCAAAAUUGGCAACAGGUUUAGUUUAUGGUUGUCGC ......((((((.....))))))......-.((..((((---(..(((...(((..((.((((----....)))).))..))).)))..)))))..))..... ( -17.30) >DroEre_CAF1 5798 92 + 1 AGCUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGA--------AAGCCAAUCACAGGCAAAAUUGGCAACAGGUUUCGAGCAGGGUUGCCGU .((.....((((.....))))(((((.....((((((..---....--------..(((.......))).....((....)))))))).....)))))))... ( -23.80) >DroYak_CAF1 5766 92 + 1 AUUUAAUUUGGCUUCAAGCCAGGCCUAAAAAGAGACUAA---AAGA--------AAGCCAAUCACAGGCAAAAUUGGCAACAGGUUUCGAGAAGGGUUGCCGU ........((((.....))))(((((.....((((((..---....--------..(((.......))).....((....)))))))).....)))))..... ( -22.80) >DroPer_CAF1 1054 90 + 1 AGAUAAUUUGGCU--AAGCCAGGCCUAAA-AGAAACUACAAGAAGA--------AAGCCAUUC--GGGCAAAAUUGGCAACAGGUUUAGCCAAGAGUGGCCAU ......(((((((--(((((..(((....-.....((......)).--------..(((....--.)))......)))....))))))))))))......... ( -26.20) >consensus AGUUAAUUUGGCUUCAAGCCAGGCCUAAA_AGAAACUAA___AAGA________AAGCCAAUC__AGGCAAAAUUGGCAACAGGUUUAGAGAAGGGUUGCCGU ........((((.....))))(((((......(((((...................(((.......))).....((....))))))).......))..))).. (-16.48 = -16.20 + -0.28)

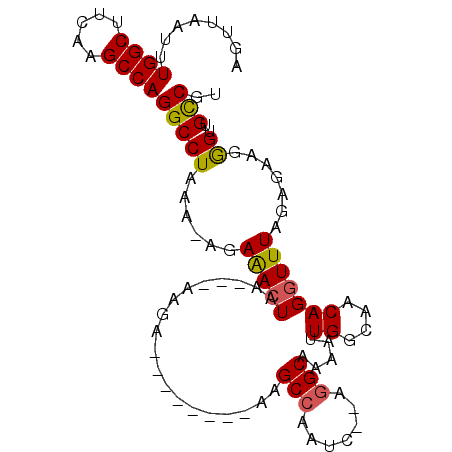
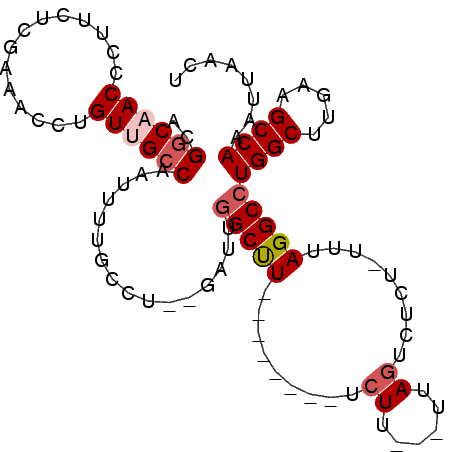
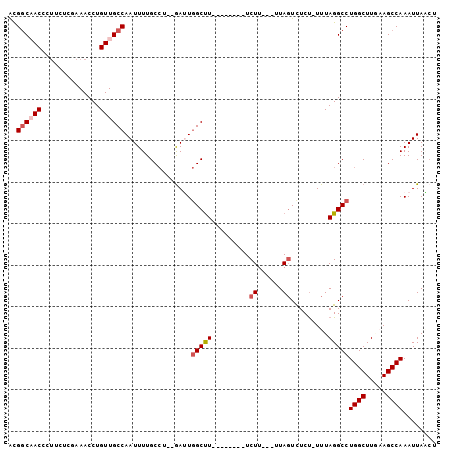
| Location | 6,335,743 – 6,335,835 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 80.08 |
| Mean single sequence MFE | -19.74 |
| Consensus MFE | -15.30 |
| Energy contribution | -15.99 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.43 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.766612 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6335743 92 - 20766785 ACGGCAACCCAGCUCGAAACCUGUUGCCAAUUUUGCCUGUGAUUGGCUU--------UCUU---UUAGUCUCUUUUUAGGCCUGGCUUGAAGCCAAAUUAACU ..((((((..............))))))......(((((.((..((((.--------....---..))))...)).))))).((((.....))))........ ( -22.74) >DroPse_CAF1 1045 90 - 1 AUGGCCACUCUUGGAUAAACCUGUUGCCAAUUUUGCCC--GAAUGGCUU--------UCUUCUUGUAGUUUCU-UUUAGGCCUGGCUU--AGCCAAAUUAUCU ..((((....((((...((....)).))))....(((.--....)))..--------................-....))))((((..--.))))........ ( -17.20) >DroGri_CAF1 5368 95 - 1 GCGACAACCAUAAACUAAACCUGUUGCCAAUUUUGC----AAUUCGCCUCAAUUUUCUCUU---CUAUUUUCC-UUUAGGCCUGGCUUAAAGCCAAAUUAACU ((((((...............)))))).........----.....((((............---.........-...)))).((((.....))))........ ( -12.52) >DroEre_CAF1 5798 92 - 1 ACGGCAACCCUGCUCGAAACCUGUUGCCAAUUUUGCCUGUGAUUGGCUU--------UCUU---UUAGUCUCUUUUUAGGCCUGGCUUGAAGCCAAAUUAGCU ..((((((..............))))))......(((((.((..((((.--------....---..))))...)).))))).((((.....))))........ ( -22.74) >DroYak_CAF1 5766 92 - 1 ACGGCAACCCUUCUCGAAACCUGUUGCCAAUUUUGCCUGUGAUUGGCUU--------UCUU---UUAGUCUCUUUUUAGGCCUGGCUUGAAGCCAAAUUAAAU ..((((((..............))))))......(((((.((..((((.--------....---..))))...)).))))).((((.....))))........ ( -22.74) >DroPer_CAF1 1054 90 - 1 AUGGCCACUCUUGGCUAAACCUGUUGCCAAUUUUGCCC--GAAUGGCUU--------UCUUCUUGUAGUUUCU-UUUAGGCCUGGCUU--AGCCAAAUUAUCU ..((((....(((((..........)))))....(((.--....)))..--------................-....))))((((..--.))))........ ( -20.50) >consensus ACGGCAACCCUUCUCGAAACCUGUUGCCAAUUUUGCCU__GAUUGGCUU________UCUU___UUAGUCUCU_UUUAGGCCUGGCUUGAAGCCAAAUUAACU ..((((((..............))))))................(((((.........((......)).........)))))((((.....))))........ (-15.30 = -15.99 + 0.70)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:22 2006