
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,333,195 – 6,333,289 |
| Length | 94 |
| Max. P | 0.877566 |

| Location | 6,333,195 – 6,333,289 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -34.90 |
| Consensus MFE | -21.84 |
| Energy contribution | -22.02 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.90 |
| SVM RNA-class probability | 0.877566 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
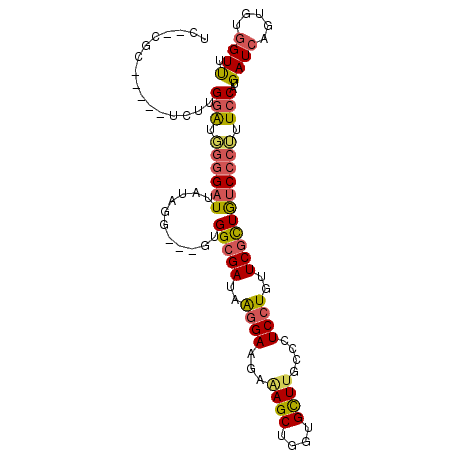
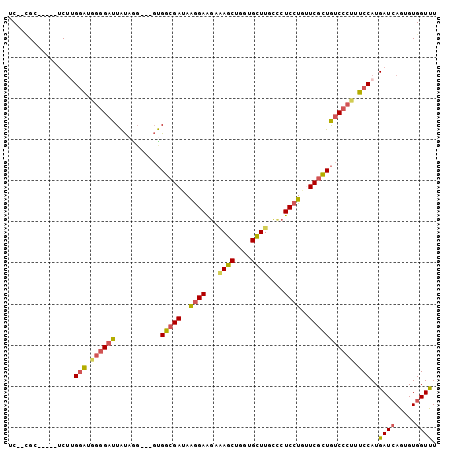
>2R_DroMel_CAF1 6333195 94 + 20766785 UC--CGC-----CCUUGGAUGGGGAUUAUAAG---GUGGCGAUAAGGAAGAAAGCUGGCGCUUGCUCUCCUGUUCGCUGUCCCUUUCCAUGAUCAGGGUGGUUU .(--(((-----(((((((.((((((......---..(((((..((((((.((((....)))).)).))))..))))))))))).)))).....)))))))... ( -40.50) >DroSim_CAF1 3226 94 + 1 UC--CGC-----UCUUGGAUGGGGAUUAUAAG---GUGGCGAUAAGGAAGAAAGCUGUUGCUUGCCCUCCUGUUCGCUGUCCCUUUCCAUGAUCAGUGUGGUUU .(--(((-----.((((((.((((((......---..(((((..((((.(.((((....))))..).))))..))))))))))).)))).....)).))))... ( -34.20) >DroEre_CAF1 3244 94 + 1 UC--CGG-----UCUUGGAUGGGGAUUAUAGG---GUGGCGAUAAGGAAGAAAGCUGGUGCUUGCCCUCCUGUUCGCUGUCCCUUUCCAUGAUCAGUGUGGUUU ..--.((-----((.((((.((((((......---..(((((..((((.(.((((....))))..).))))..))))))))))).)))).)))).......... ( -35.10) >DroYak_CAF1 3258 94 + 1 UC--UGC-----UCUAGGAUGGGGAUUAUAGG---GUGGCGAUAAGGAAGAAAGCUGGUGCUUGCCCUCCUGUUCGCUGUCCCUUUCCAUGAUCAGUGUGGUUU .(--..(-----.((..(((.((((....(((---(((((((..((((.(.((((....))))..).))))..)))))).)))).))).).))))).)..)... ( -31.90) >DroMoj_CAF1 3222 102 + 1 UCUCUGGAGUGUACAUGGAUG--GAUUACUUUGCUCUGGCGACAGGGAGCAGAGCUGGCGUUUAUUCUCCUGCUCGUUGUCUCGCUCCAUGAUGAGAGUGGUCU (((.((.......)).))).(--(((((((((...(((....)))(((((.((((.((.(......).)).))))(......))))))......)))))))))) ( -31.90) >DroAna_CAF1 4478 94 + 1 UC--GGG-----UCUGGUGUUGGGCUUGUAGG---GCGGGGAAAGCGAGGACAGCUGGUGCUUGUUCUCCUGCUCGCUAUCCCGCUCCAUGAUCAGAGUGGUCU .(--.(.-----((((((..(((((..(...(---(((((...((.(((((((((....)).))))))))).))))))..)..)).)))..)))))).).)... ( -35.80) >consensus UC__CGC_____UCUUGGAUGGGGAUUAUAGG___GUGGCGAUAAGGAAGAAAGCUGGUGCUUGCCCUCCUGUUCGCUGUCCCUUUCCAUGAUCAGUGUGGUUU ................(((.((((((...........(((((..((((...((((....))))....))))..))))))))))).)))..((((.....)))). (-21.84 = -22.02 + 0.17)

| Location | 6,333,195 – 6,333,289 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 78.36 |
| Mean single sequence MFE | -28.60 |
| Consensus MFE | -11.59 |
| Energy contribution | -13.93 |
| Covariance contribution | 2.34 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.41 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.823387 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
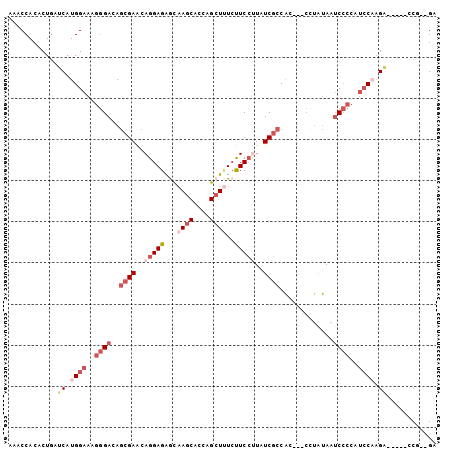
>2R_DroMel_CAF1 6333195 94 - 20766785 AAACCACCCUGAUCAUGGAAAGGGACAGCGAACAGGAGAGCAAGCGCCAGCUUUCUUCCUUAUCGCCAC---CUUAUAAUCCCCAUCCAAGG-----GCG--GA ...((.((((.....((((..((((..((((..((((.((.((((....)))).))))))..))))...---.......))))..)))))))-----).)--). ( -31.20) >DroSim_CAF1 3226 94 - 1 AAACCACACUGAUCAUGGAAAGGGACAGCGAACAGGAGGGCAAGCAACAGCUUUCUUCCUUAUCGCCAC---CUUAUAAUCCCCAUCCAAGA-----GCG--GA ........(((.((.((((..((((..((((..(((((((.((((....)))))))))))..))))...---.......))))..)))).))-----.))--). ( -30.40) >DroEre_CAF1 3244 94 - 1 AAACCACACUGAUCAUGGAAAGGGACAGCGAACAGGAGGGCAAGCACCAGCUUUCUUCCUUAUCGCCAC---CCUAUAAUCCCCAUCCAAGA-----CCG--GA ........(((.((.((((..((((..((((..(((((((.((((....)))))))))))..))))...---.......))))..)))).))-----.))--). ( -30.40) >DroYak_CAF1 3258 94 - 1 AAACCACACUGAUCAUGGAAAGGGACAGCGAACAGGAGGGCAAGCACCAGCUUUCUUCCUUAUCGCCAC---CCUAUAAUCCCCAUCCUAGA-----GCA--GA ........(((.((..(((..((((..((((..(((((((.((((....)))))))))))..))))...---.......))))..)))..))-----.))--). ( -28.70) >DroMoj_CAF1 3222 102 - 1 AGACCACUCUCAUCAUGGAGCGAGACAACGAGCAGGAGAAUAAACGCCAGCUCUGCUCCCUGUCGCCAGAGCAAAGUAAUC--CAUCCAUGUACACUCCAGAGA ......((((...((((((..((((((..((((((.((............))))))))..))))((....)).......))--..))))))........)))). ( -25.20) >DroAna_CAF1 4478 94 - 1 AGACCACUCUGAUCAUGGAGCGGGAUAGCGAGCAGGAGAACAAGCACCAGCUGUCCUCGCUUUCCCCGC---CCUACAAGCCCAACACCAGA-----CCC--GA .......((((....(((.((((((.((((((.....((...(((....))).)))))))).)))).(.---....)..)))))....))))-----...--.. ( -25.70) >consensus AAACCACACUGAUCAUGGAAAGGGACAGCGAACAGGAGAGCAAGCACCAGCUUUCUUCCUUAUCGCCAC___CCUAUAAUCCCCAUCCAAGA_____CCG__GA ............((.((((..((((..((((..(((((...((((....))))..)))))..)))).............))))..)))).))............ (-11.59 = -13.93 + 2.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:19 2006