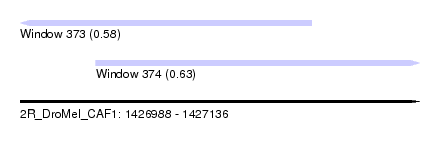
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 1,426,988 – 1,427,136 |
| Length | 148 |
| Max. P | 0.626814 |

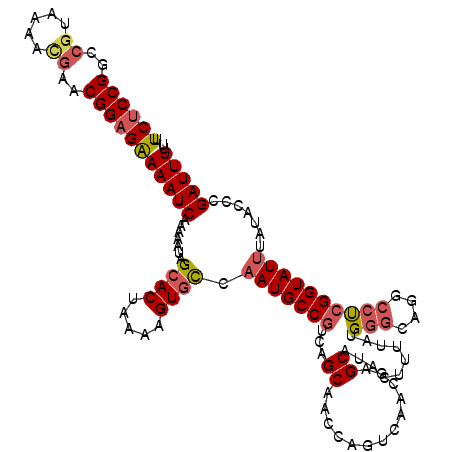
| Location | 1,426,988 – 1,427,096 |
|---|---|
| Length | 108 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.45 |
| Mean single sequence MFE | -36.82 |
| Consensus MFE | -21.48 |
| Energy contribution | -23.08 |
| Covariance contribution | 1.60 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.581886 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1426988 108 - 20766785 UGCUCGUUGACUGGUUGCUGACGGCAUUGGAACUUUUAGUGCUAUUUUUGAUUUUCUCCGUUCGUUUUACGGCCGGAGAACAGCGGGAGGAAUA------------CAGGCAAAUUAACA (((..((...((..((((((..(((((((((...)))))))))..........(((((((..((.....))..)))))))))))))..))...)------------)..)))........ ( -35.30) >DroSec_CAF1 45428 108 - 1 UGCUCGUUGACUGGUUGCUGACGGCAUUGGCACUUUUAGUGCUAUUUUUGAUUUUCUCCGUUCGUUUUACGGCCGGAGAACGGCGGGAGGAGUA------------CGGGCAAAUUAACA (((((((...((..((((((.(((...((((((.....))))))...)))...(((((((..((.....))..)))))))))))))..))...)------------))))))........ ( -40.50) >DroSim_CAF1 50485 108 - 1 UGCUCGUUGACUGGUUGCUGACGGCAUUGGCACUUUUAGUGCUAUUUUUGAUUUUCUCCGUUCGUUUUACGGCCGGAGAACGGCGGGAGGAGUA------------CAGGCAAAUUAACA (((..((...((..((((((.(((...((((((.....))))))...)))...(((((((..((.....))..)))))))))))))..))...)------------)..)))........ ( -35.60) >DroEre_CAF1 43501 118 - 1 UGCUCGCUGACUGGUUGCCAUCGGCAUAGGCACUUUUAGUGCUAUUUUUGAUUUUCUCCGUUGGUUUCAGGGCCGGCGAGCAGCGGGG--UAUGCUCGCACUCGCACAAGCGAAUUAACA (((((((((.((((..((((.(((....(((((.....)))))......(.....).))).)))).))))...)))))))))(((((.--....)))))..((((....))))....... ( -45.20) >DroYak_CAF1 48089 99 - 1 GGCU---------GUUGCUGACGGCAUUGACACUUUUAGUGCUAUUUUUGAUUUCCUCCGUUCAUUUUACGGCCGGAGAACAGCGGGGGGAAUG------------CAGGUUAAUUAACA ....---------((((.((((((((((((.....))))))))....(((((((((((((((..((((.......))))..)))))))))))).------------)))))))..)))). ( -27.50) >consensus UGCUCGUUGACUGGUUGCUGACGGCAUUGGCACUUUUAGUGCUAUUUUUGAUUUUCUCCGUUCGUUUUACGGCCGGAGAACAGCGGGAGGAAUA____________CAGGCAAAUUAACA ..(((((((.............((((((((.....))))))))..........(((((((..((.....))..))))))))))))))................................. (-21.48 = -23.08 + 1.60)

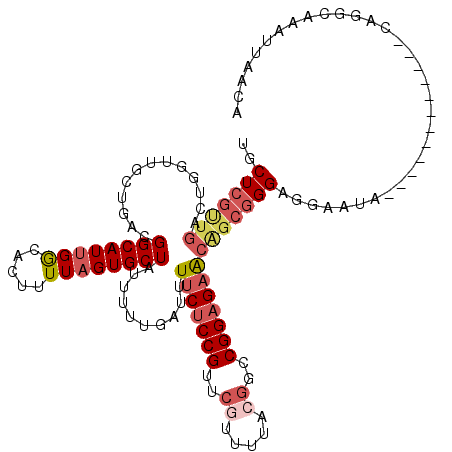
| Location | 1,427,016 – 1,427,136 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 86.57 |
| Mean single sequence MFE | -34.01 |
| Consensus MFE | -20.09 |
| Energy contribution | -21.14 |
| Covariance contribution | 1.05 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.59 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.626814 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 1427016 120 + 20766785 UUCUCCGGCCGUAAAACGAACGGAGAAAAUCAAAAAUAGCACUAAAAGUUCCAAUGCCGUCAGCAACCAGUCAACGAGCAUACUUUUAUGGGCAGGCCUCGGUAUUUAUACCCGAUUUUU .((((((..((.....))..))))))(((((.......((..(((((((....(((((((..((.....))..))).)))))))))))...)).......((((....)))).))))).. ( -32.10) >DroSec_CAF1 45456 120 + 1 UUCUCCGGCCGUAAAACGAACGGAGAAAAUCAAAAAUAGCACUAAAAGUGCCAAUGCCGUCAGCAACCAGUCAACGAGCAUACUUUUAUGGGCAGGCCUCGGUAUUUUUAGGCGAUUUUU (((((((..((.....))..)))))))....((((((.((.(((((((((((.(((((((..((.....))..))).))))........(((....))).))))))))))))).)))))) ( -41.60) >DroSim_CAF1 50513 120 + 1 UUCUCCGGCCGUAAAACGAACGGAGAAAAUCAAAAAUAGCACUAAAAGUGCCAAUGCCGUCAGCAACCAGUCAACGAGCAUACUUUUAUGGGCAGGCCUCGGUAUUUAUACCCGAUUUUU .((((((..((.....))..))))))(((((.......((((.....)))).(((((((...((..(((......(((....)))...)))....))..))))))).......))))).. ( -32.20) >DroEre_CAF1 43539 119 + 1 CUCGCCGGCCCUGAAACCAACGGAGAAAAUCAAAAAUAGCACUAAAAGUGCCUAUGCCGAUGGCAACCAGUCAGCGAGCAUACUUUUAUGGGCAGG-CCGGGUAUUUAUACCCGAUUUUU ...(((.(((((((((......................((((.....)))).(((((((.((((.....)))).)).))))).))))).)))).))-)((((((....))))))...... ( -39.50) >DroYak_CAF1 48117 104 + 1 UUCUCCGGCCGUAAAAUGAACGGAGGAAAUCAAAAAUAGCACUAAAAGUGUCAAUGCCGUCAGCAAC---------AGCCCAAUUCAAUG-------CUCGGUAUUUAUACCCGAUUUUU ..(((((..(((...)))..)))))((((((.......((((.....)))).(((((((..((((..---------............))-------))))))))).......)))))). ( -24.64) >consensus UUCUCCGGCCGUAAAACGAACGGAGAAAAUCAAAAAUAGCACUAAAAGUGCCAAUGCCGUCAGCAACCAGUCAACGAGCAUACUUUUAUGGGCAGGCCUCGGUAUUUAUACCCGAUUUUU .((((((..((.....))..))))))(((((.......((((.....)))).(((((((...((.............))..........(((....)))))))))).......))))).. (-20.09 = -21.14 + 1.05)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:30:52 2006