| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,311,579 – 6,311,679 |
| Length | 100 |
| Max. P | 0.671678 |

| Location | 6,311,579 – 6,311,679 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 80.37 |
| Mean single sequence MFE | -30.20 |
| Consensus MFE | -18.97 |
| Energy contribution | -19.78 |
| Covariance contribution | 0.81 |
| Combinations/Pair | 1.32 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.29 |
| SVM RNA-class probability | 0.671678 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6311579 100 - 20766785 AGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCUCACAUCGUCCCCUAAUCGAGCCCUUU--UCUCCAGGGACACAAACAGAGGGCGGAG--CGGCCAUAA .......((((....).((((........(((((.((......(((((......(((......--.)))..)))))......)).)))))...--))))))).. ( -24.00) >DroPse_CAF1 46376 87 - 1 AGUCAUAAUGGGCAACAGCUGAUAACACACGCCCCCCCACAA--C------------CUCUUUAUUCUUC-GAGAGAGAG--AGAGGCGGAAACGGGGCUAUAA .......((.(((....))).)).......(((((.......--(------------((((((.(((((.-..)))))))--))))).(....))))))..... ( -26.90) >DroSec_CAF1 61775 99 - 1 AGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCGCCCAUCGCCCCU-AAUCGAGCCCUUU--UCUCCAGGGACACAAACAGGGGGCGGAA--CGGCCAUAA .(((....((((.....((((......)).)).....))))((((((((-.......((((..--.....)))).........))))))))..--.)))..... ( -28.79) >DroSim_CAF1 38648 100 - 1 AGUCAUAAUGGGAAACAGCUGAUAGUACAAGCCCCCGCCCAUCGCCCCCAAAUCGAGCCCUUU--UCUCCAGGGACACAAACAGGGGGCGGAA--CGGCCAUAA .(((....((((.....(((.........))).....))))((((((((........((((..--.....)))).........))))))))..--.)))..... ( -32.13) >DroEre_CAF1 56865 99 - 1 AGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCUCCCACCGCCCCCUAAUCGAGCCC-UU--UCUCCAGGGACACAGAAAGGGGGCGGGG--CGGCCAUAA .(((....(((((....((((......)).))....)))))(((((((((..((...(((-(.--.....)))).....)).)))))))))))--)........ ( -37.90) >DroYak_CAF1 83251 99 - 1 AGUCAUAAUGGAAAACAGCUGAUAGUACACGCCCCCUUCCACUGCCCCCUAAUCGAGCCU-UU--UCUCUGGGGACACAAACAGGGGGCGGGG--CAGCCAUAA .(((....(((((....((((......)).))....)))))(((((((((....(((...-..--.)))((....)).....)))))))))))--)........ ( -31.50) >consensus AGUCAUAAUGGGAAACAGCUGAUAGUACACGCCCCCUCCCAUCGCCCCCUAAUCGAGCCCUUU__UCUCCAGGGACACAAACAGGGGGCGGAA__CGGCCAUAA .(((....((((.....((((......)).)).....))))((((((((........(((...........))).........)))))))).....)))..... (-18.97 = -19.78 + 0.81)

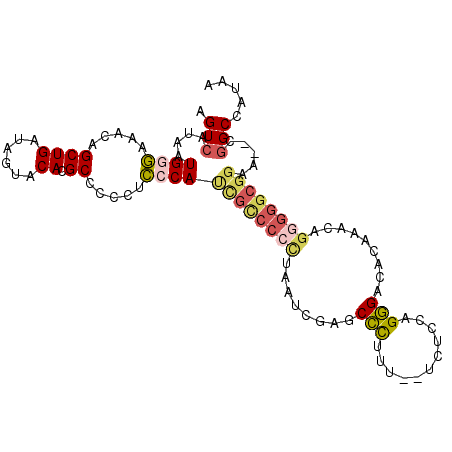
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:12 2006