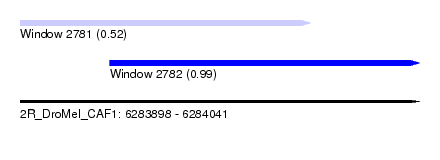
| Sequence ID | 2R_DroMel_CAF1 |
|---|---|
| Location | 6,283,898 – 6,284,041 |
| Length | 143 |
| Max. P | 0.991396 |

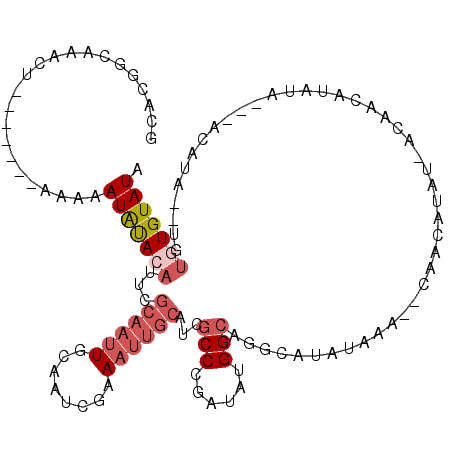
| Location | 6,283,898 – 6,284,002 |
|---|---|
| Length | 104 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.90 |
| Mean single sequence MFE | -24.46 |
| Consensus MFE | -7.30 |
| Energy contribution | -8.58 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.30 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.518446 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6283898 104 + 20766785 GCCGCCAACCAUUUUAGCUUUGACC--------AAGUUACGCACGGCAAACU-------AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAAA- (((((((....((((((.((((.((--------..(.....)..))))))))-------))))..........((...((((((....))))))..))......)))).))).......- ( -25.10) >DroSec_CAF1 26512 101 + 1 GC---CAACCAUUUUAGCUUUGACC--------AAGUUACGCACGGCAAACU-------AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAUA- ((---(.....((((((.((((.((--------..(.....)..))))))))-------))))..........((((((........))))))...(((......))).))).......- ( -22.50) >DroEre_CAF1 27447 109 + 1 GC---CAACCAUUUUAGCUUUGACC--------AAGUUACGCACUGCAAACUAAAAACUAAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAGUA ((---(.....((((((.((((...--------.(((.....))).)))))))))).................((((((........))))))...(((......))).)))........ ( -21.60) >DroYak_CAF1 29774 101 + 1 GC---CAACCAUUUUAGCUUUGACC--------AAGUUACGCACGGCAAACU-------AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAGA- ((---(.....((((((.((((.((--------..(.....)..))))))))-------))))..........((((((........))))))...(((......))).))).......- ( -22.50) >DroAna_CAF1 28308 101 + 1 GU---CAAUCACUUCAGCUGCGCCCGAAUGGCUGAGUGGCGAAUGGCGGCCC-------AAAAAUGCACAAUUGCAAGUGCAGUGGCA-------GGCCCGAUAUGGAUUGGA-AUAAA- .(---(((((...((.(((.(((((..........).))))...)))((((.-------.....(((((........)))))......-------)))).))....)))))).-.....- ( -30.60) >consensus GC___CAACCAUUUUAGCUUUGACC________AAGUUACGCACGGCAAACU_______AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAAA_ ................((((.....................................................((((((........))))))...(((......)))))))........ ( -7.30 = -8.58 + 1.28)

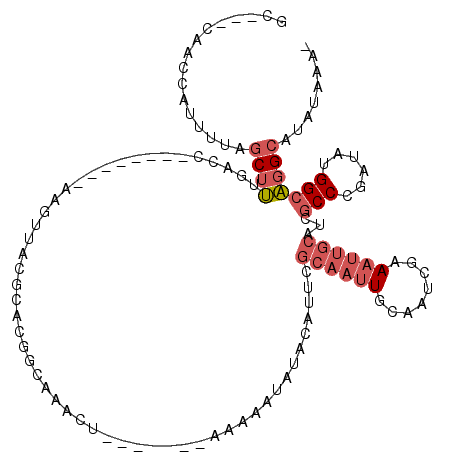
| Location | 6,283,930 – 6,284,041 |
|---|---|
| Length | 111 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.49 |
| Mean single sequence MFE | -22.22 |
| Consensus MFE | -9.10 |
| Energy contribution | -10.58 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.36 |
| Structure conservation index | 0.41 |
| SVM decision value | 2.27 |
| SVM RNA-class probability | 0.991396 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>2R_DroMel_CAF1 6283930 111 + 20766785 GCACGGCAAACU-------AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAAA--CAAAAAAAAAAAACAUACACAUACAUAAGUGUGUGUAUA .....((.....-------..............((((((........))))))...(((......)))..)).......--..............((((((((((....)))))))))). ( -24.50) >DroSec_CAF1 26541 92 + 1 GCACGGCAAACU-------AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAUA--CAACAA------ACAUAUA------UA------UGUAU- ............-------..............((((((........))))))...(((......)))..(((((((((--......------...))))------))------)))..- ( -16.40) >DroEre_CAF1 27476 111 + 1 GCACUGCAAACUAAAAACUAAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAGUAUAAACAUAU-ACAGCAU------ACAUA--UGUGUGUAUA ((.((((..........................((((((........))))))(((....)))...)))))).....(((((....)))-)).((((------((...--.))))))... ( -24.30) >DroYak_CAF1 29803 107 + 1 GCACGGCAAACU-------AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAGA--CAAUAUAU-ACAAUAUAUACA-ACAUA--UGUGUGUAUA .....((.....-------..............((((((........))))))...(((......)))..))(((((..--...)))))-...(((((((((-.....--))))))))). ( -20.00) >DroAna_CAF1 28345 103 + 1 GAAUGGCGGCCC-------AAAAAUGCACAAUUGCAAGUGCAGUGGCA-------GGCCCGAUAUGGAUUGGA-AUAAA--CAAUAUAGUAUAGUAUAGUACUAUAUGGUGCUCUGUAUG ..((((.(((((-------.....(((...(((((....))))).)))-------...(((((....))))).-.....--..(((((((((......))))))))))).)))))))... ( -25.90) >consensus GCACGGCAAACU_______AAAAAUAUACAUUCGCAAUUGCAAUCGAAAUUGCAUCGCCCGAUAUGGCAGGCAUAUAAA__CAACAUAU_ACAACAUAUA___ACAUA__UGUGUGUAUA .......................(((((((...((((((........))))))...(((......)))............................................))))))). ( -9.10 = -10.58 + 1.48)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 10:10:09 2006